Method for regulating and controlling self-assembly of nucleic acid nanostructure based on DNA stem-loop conformation transformation
A nucleic acid nanometer and nanostructure technology, applied in the field of DNA nanometers, can solve problems such as restrictions on wider application and lack, and achieve the effect of flexible regulation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
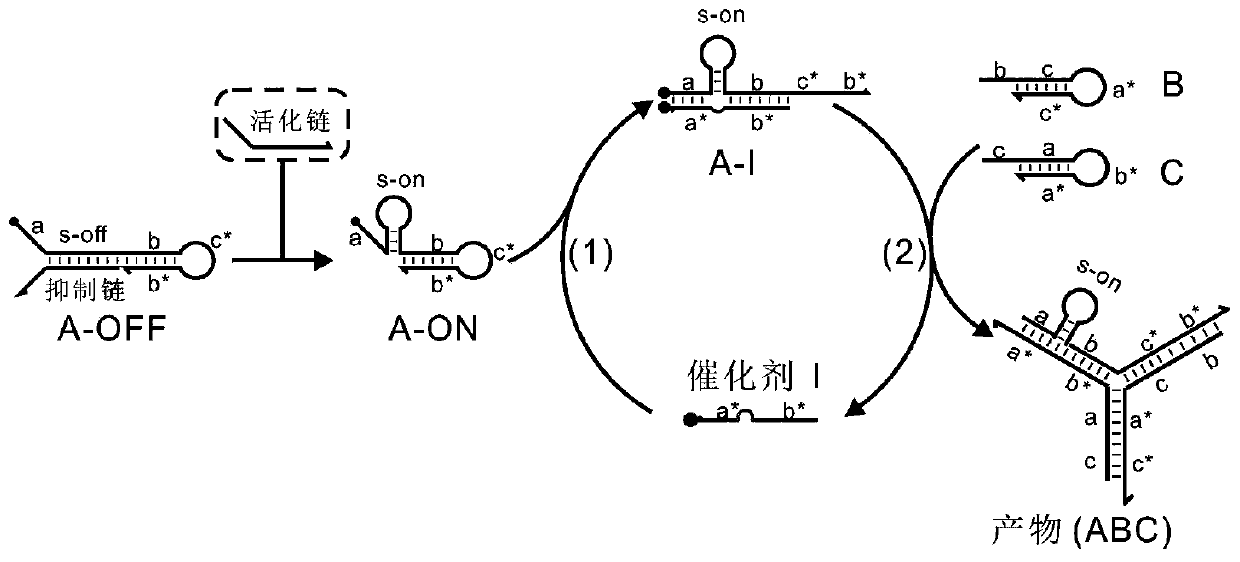
[0065] (1) Design the sequences of DNA strands A, B, C, and I required for assembling the Y-shaped structure, wherein the 5' to 3' ends of the sequence A are successively a, s, b, c*, b* regions, at 5 'modified FAM fluorescent group; the 5' to 3' of sequence B are b, c, a*, c* regions in sequence; the 5' to 3' ends of sequence C are c, a, b*, a* regions in sequence, Dabcyl quenching group is modified at 3'; the 5' to 3' ends of sequence I are sequentially b*, t, and a* regions; wherein a, b, and c regions are complementary to the sequences of a*, b*, and c* regions, respectively For pairing, the S region is the stem-loop region, and the main region is inserted into a 6nt protection region to stabilize the key structure in the reaction pathway.
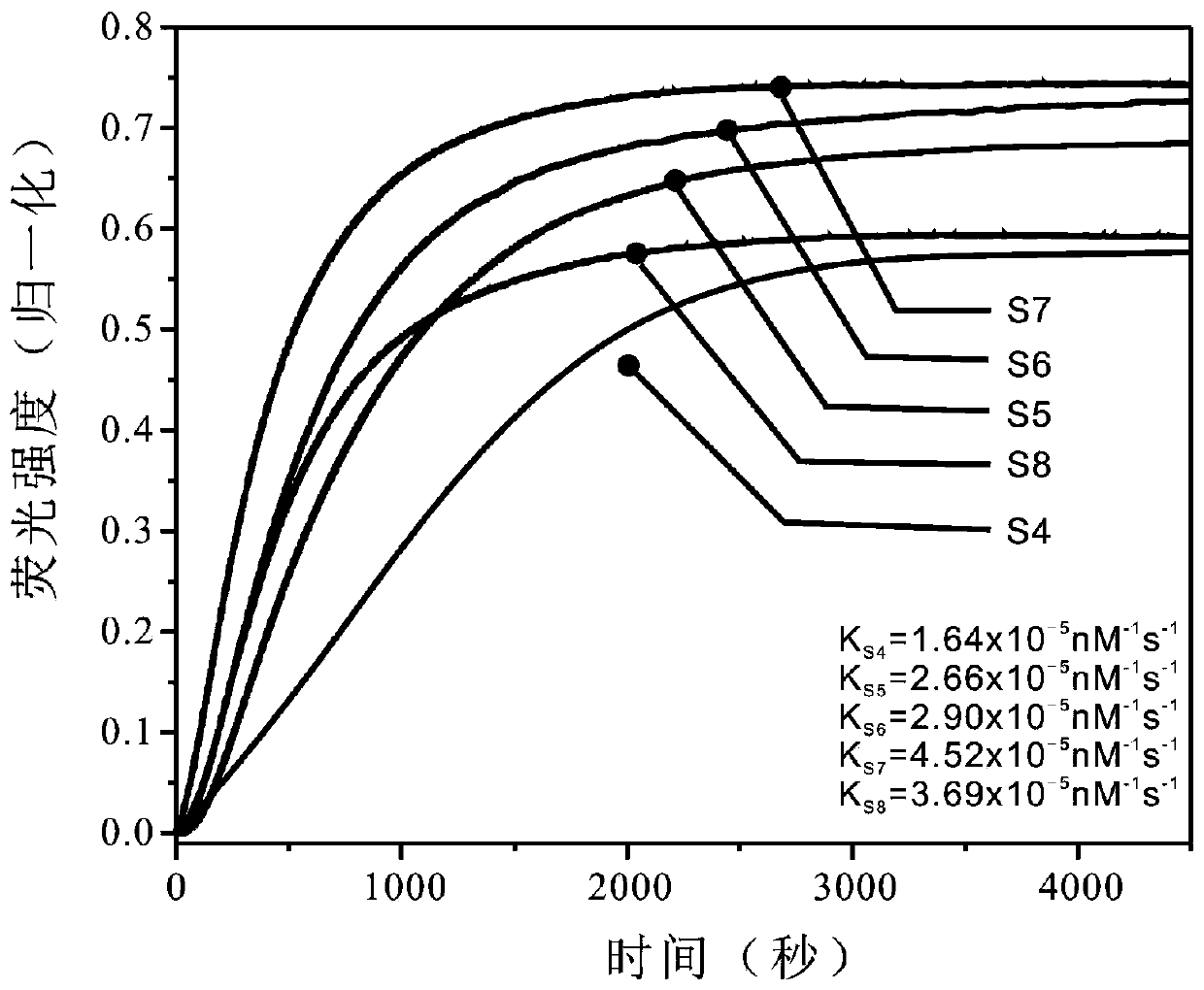
[0066] (2) Design the sequences of the stems of the stem-loop structure in chain A, the lengths of which are 4, 5, 6, 7, and 8 nt, respectively.
[0067] (3) At 25°C, the nucleic acid sequences A, B, C, and I were added to TE / Mg in a ...
Embodiment 2
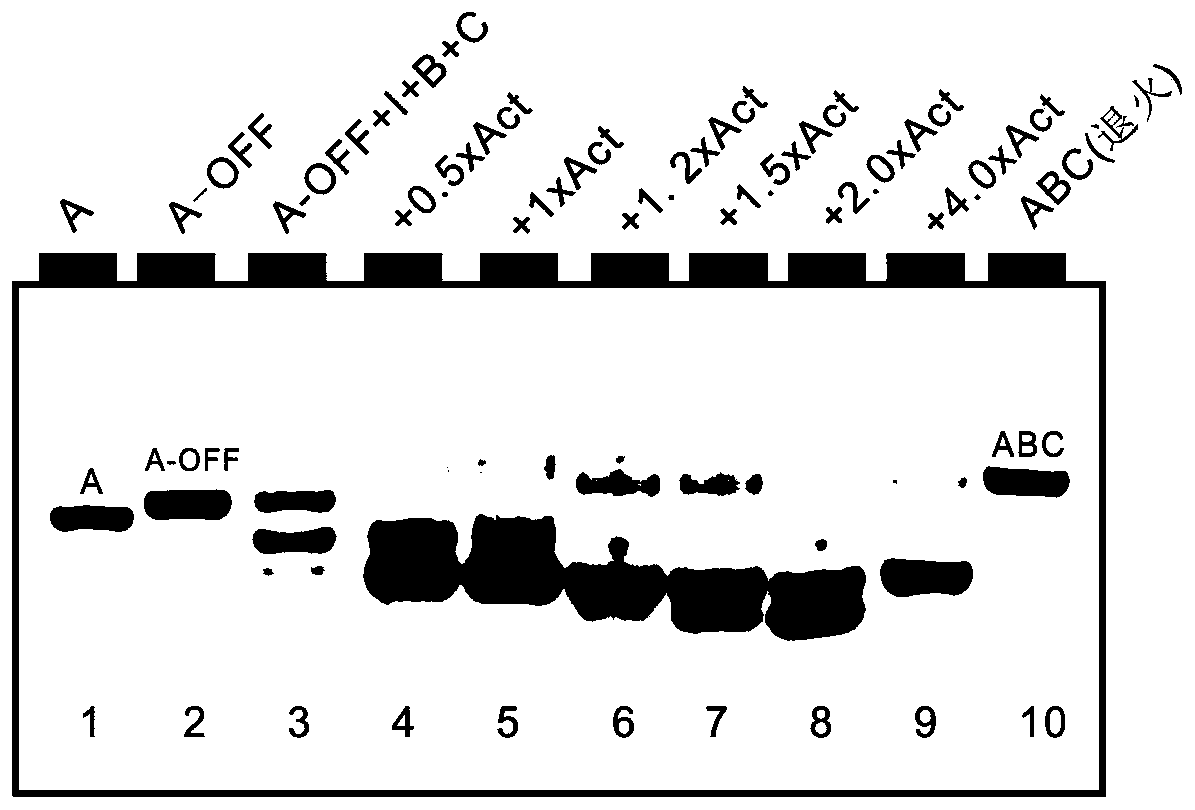
[0069] (1) At 25°C, the A chain in the S-Off state and the chains B, C, and I were mixed in TE / Mg at a molar ratio of 1:1.2:1.5:1 2+ The buffer system is mixed well. At this time, because the switch has a rigid double-chain structure, which "separates" the branch migration end from the toehold end, the chain replacement reaction is difficult to proceed, so no Y-shaped structure is formed.
[0070] (2) Add 2.5nM, 5nM, 6nM, 7.5nM, 10nM and 40nM activated strands to the above buffer solution respectively. Due to the chain displacement reaction between the activated strand and the inhibitory strand, a stem-loop structure is formed, and the branch migration end and toehold The end distance is shortened, the hairpin A structure is opened by chain I, and then B, C and A pair with each other to form a Y-shaped nanostructure of A-B-C. The minimum concentration of the activated chain added was 2.5nM (0.1x), which indicated that the conformational transformation of the stem-loop switch w...
Embodiment 3
[0072] Because a stem-loop switch can only regulate whether the Y-shaped nanostructure is formed, but cannot finely regulate the intermediate assembly process. In this experiment, three stem-loop switches with different sequences were used to implement multi-step controllability on Y-shaped nanostructures.
[0073] (1) Sequence design
[0074] Add a stem-loop switch to the toehold and branch migration regions of the A, B, and C strands respectively, and design the sequences of three stem-loop structures, in which the length of the stem is 7nt, and the loop is 15nt; design the inhibitory chain and the activation chain corresponding to the stem-loop , and its toehold length is 7nt.
[0075] (2) Step-by-step regulation
[0076] At 25°C, the A, B, C chains and I chains in the S-Off state were mixed in TE / Mg at a molar ratio of 1:1.2:1.5:1. 2+ Mix the buffer system. Since there are three stem-loop switches in the system, different structures can be produced by adding different ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com