Spatial indexing of genetic material and library preparation using hydrogel beads and flow cells
A technology of hydrogel beads, genetic material, applied in specific-purpose bioreactors/fermenters, libraries, chemical libraries, etc., can solve problems such as increasing the complexity of SBS methods, achieve simplified sequence reconstruction, improve efficiency, eliminate The effect of cumbersome barcode steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
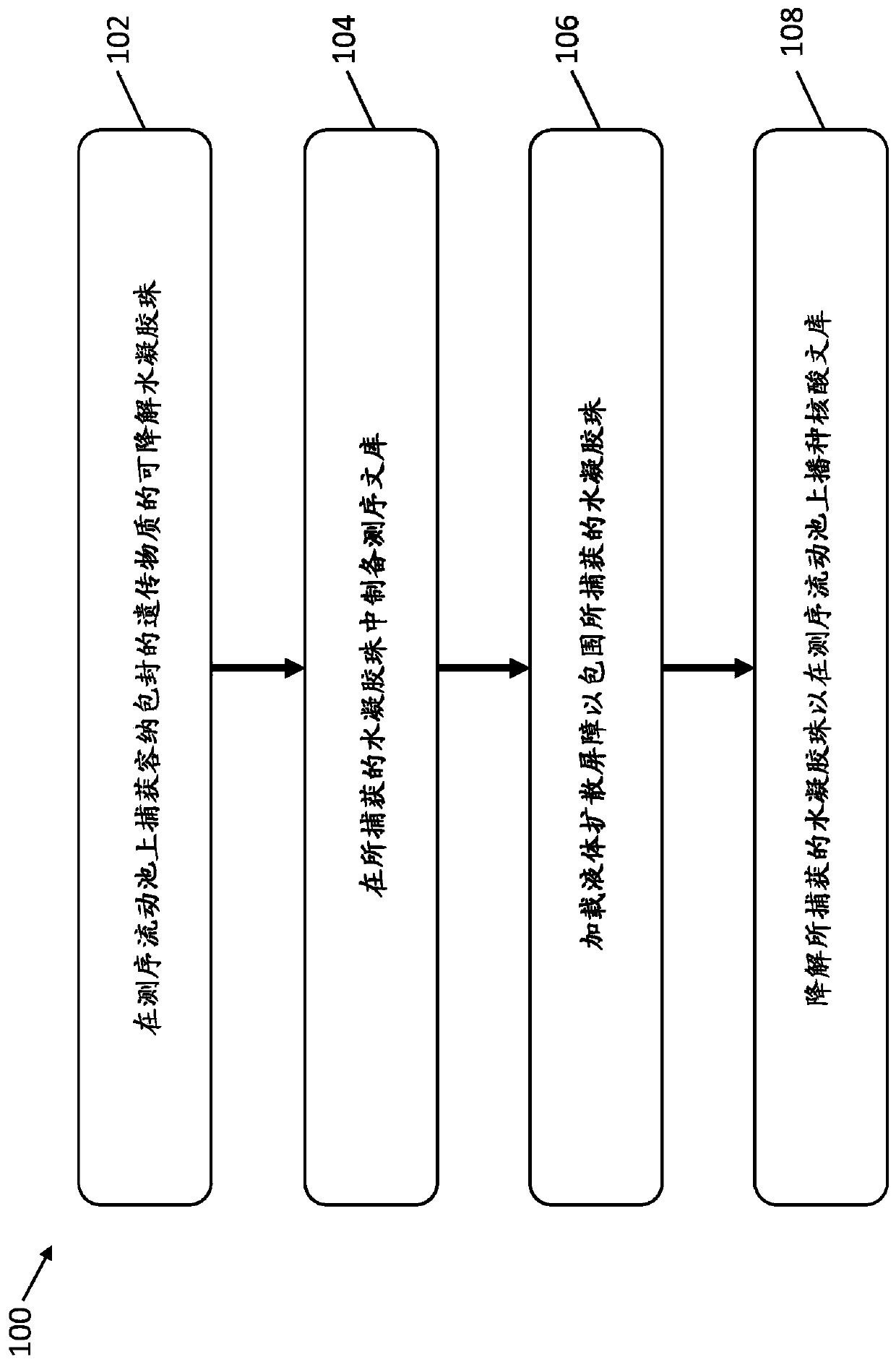
[0305] Flow Cell Capture of Degradable Hydrogel Beads Holding Encapsulated Nucleic Acid Molecules
[0306] This example illustrates the preparation of hydrogel beads encapsulating nucleic acid molecules using a manual vortex and a microfluidic droplet generator.
[0307] The pore size of the hydrogel beads is chosen to allow diffusion of enzymes, chemicals, and smaller sized primers (300bps), allowing genomic DNA and resulting DNA libraries to remain in the gel beads during the process.
[0308] A 12% hydrogel solution was prepared from 40% acrylamide / bis 19:1 (BioRad #161-0144) diluted with deionized water. Prepare a 40% (w / v) acrylamide / N,N'bis(acryloyl)cystamine (BACy) (19:1) monomer stock solution (3.8 g acrylamide, 0.2 g BACy and 6 mL double distilled (dd) HO ). The mixture was brought to a final volume of 10 mL. To prepare the solution, dissolve acrylamide in 6 mL of ddHO and use the resulting solution to dissolve BACy. Dissolution of the monomer is endothermic, so ...
Embodiment 2
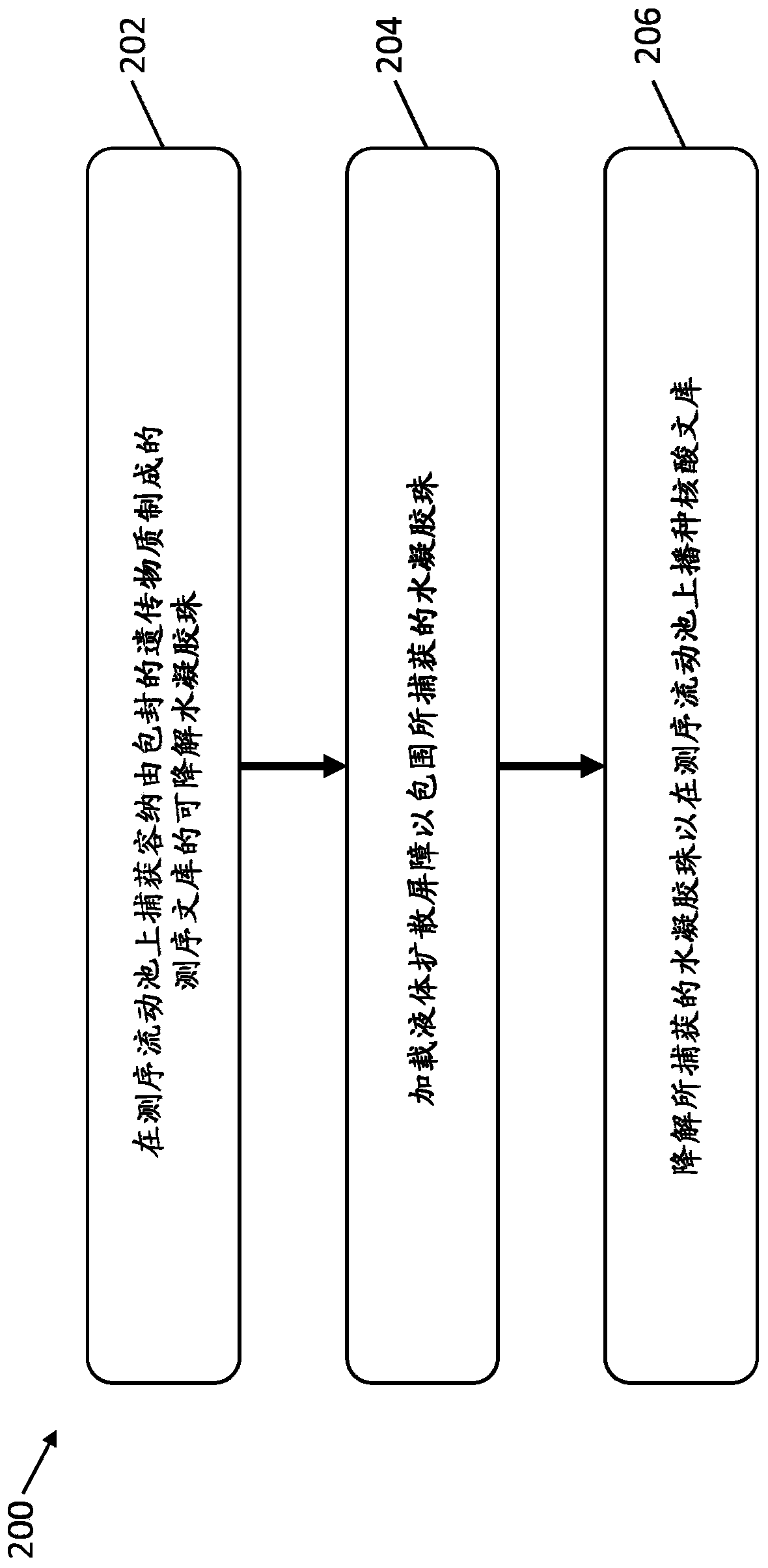
[0317] In-bead sequencing library preparation and flow cell seeding
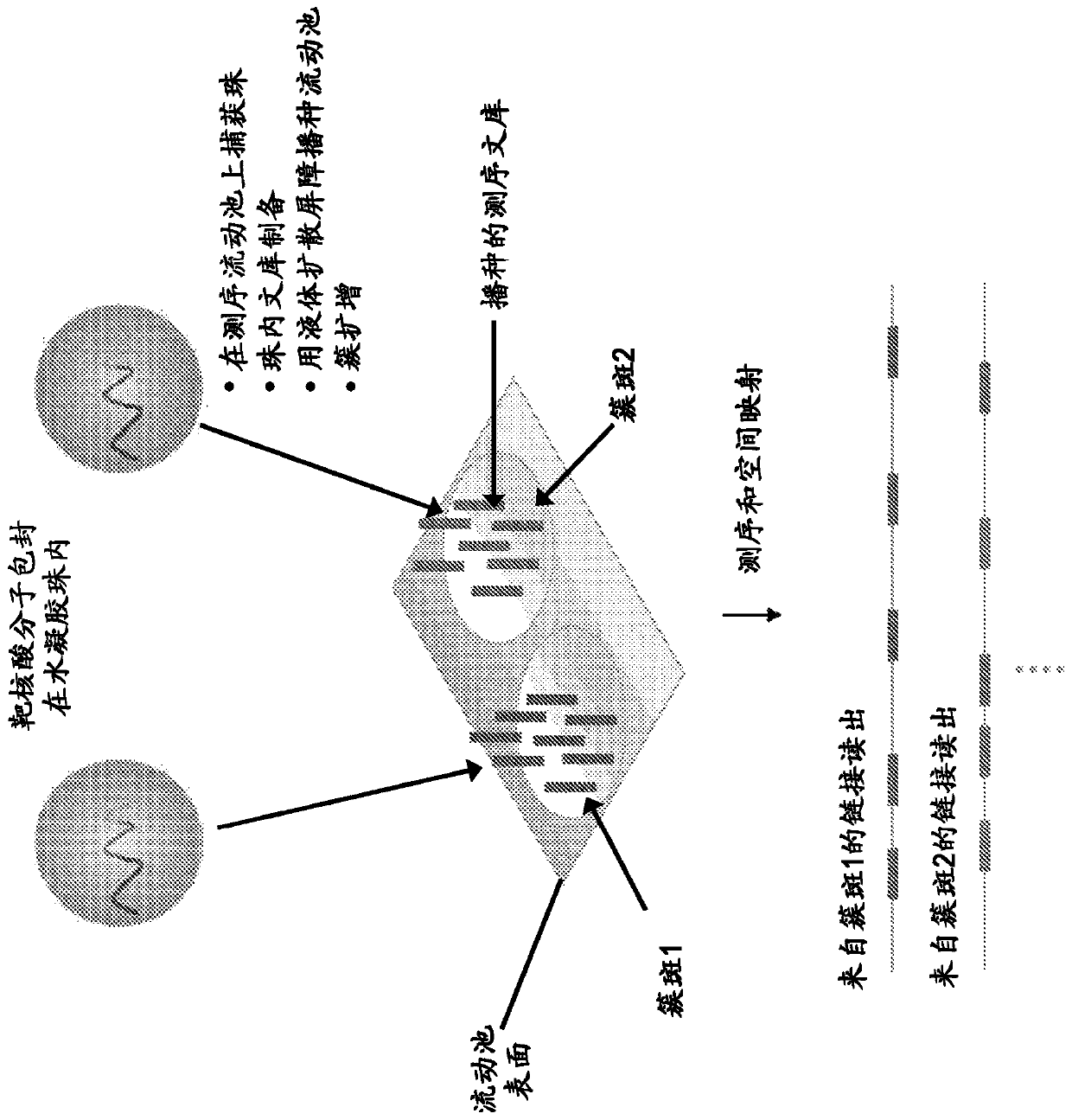
[0318] This example illustrates the preparation of a sequencing library from genomic DNA held in hydrogel beads captured on a sequencing flow cell, and the subsequent seeding of the prepared nucleic acid library on the flow cell.
[0319] As described in Example 1, in the MiSeq TM Hydrogel beads containing the encapsulated genomic DNA fragments are captured on the surface of the flow cell. A solution containing enzymes and reagents for DNA library preparation from genomic DNA in beads captured on the sequencing flow cell was flowed onto the flow cell to prepare the DNA library in hydrogel beads ( Figure 4A with 4B ). The pore size of the captured hydrogel beads allows diffusion of enzymes, chemicals, and smaller sized primers (300bps). Mineral oil is then loaded onto the sequencing flow cell to fill the voids between the beads and surround the captured hydrogel beads containing the DNA library. Then th...
Embodiment 3
[0322] Sequencing Nucleic Acids Using Hydrogel Beads
[0323] This example presents the results of a sequencing assay using hydrogel beads to spatially index nucleic acid molecules on the surface of a flow cell.
[0324] Three different workflows were determined for sequencing long DNA molecules (see Figures 5A-5C ). Two different DNA fragment lengths (~100 kb and ~10 kb) were tested, along with an in-bead MDA step prior to sequencing library preparation to amplify genomic DNA in hydrogel beads.
[0325] Hydrogel beads containing encapsulated genomic DNA fragments of ~100 kb or ~10 kb were generated as described in Example 1 . For each fragment length, in-bead MDA was assessed prior to library generation.
[0326] Load the resulting hydrogel beads into the MiSeq TM Sequencing on a flow cell and using Nextera TM (Illumina, SanDiego, CA) reagents and protocol to generate sequencing libraries by tagging reactions. Mineral oil is then loaded into the flow cell to surround t...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com