mecA GENE AMPLIFICATION PRIMER PAIR, mecA GENE DETECTION KIT AND mecA GENE DETECTION METHOD
A detection method and primer pair technology, applied in biochemical equipment and methods, microbiological determination/inspection, DNA/RNA fragments, etc., can solve problems such as infection of susceptible hosts, and achieve high sensitivity and high precision results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0164] (Example 1) primary screening
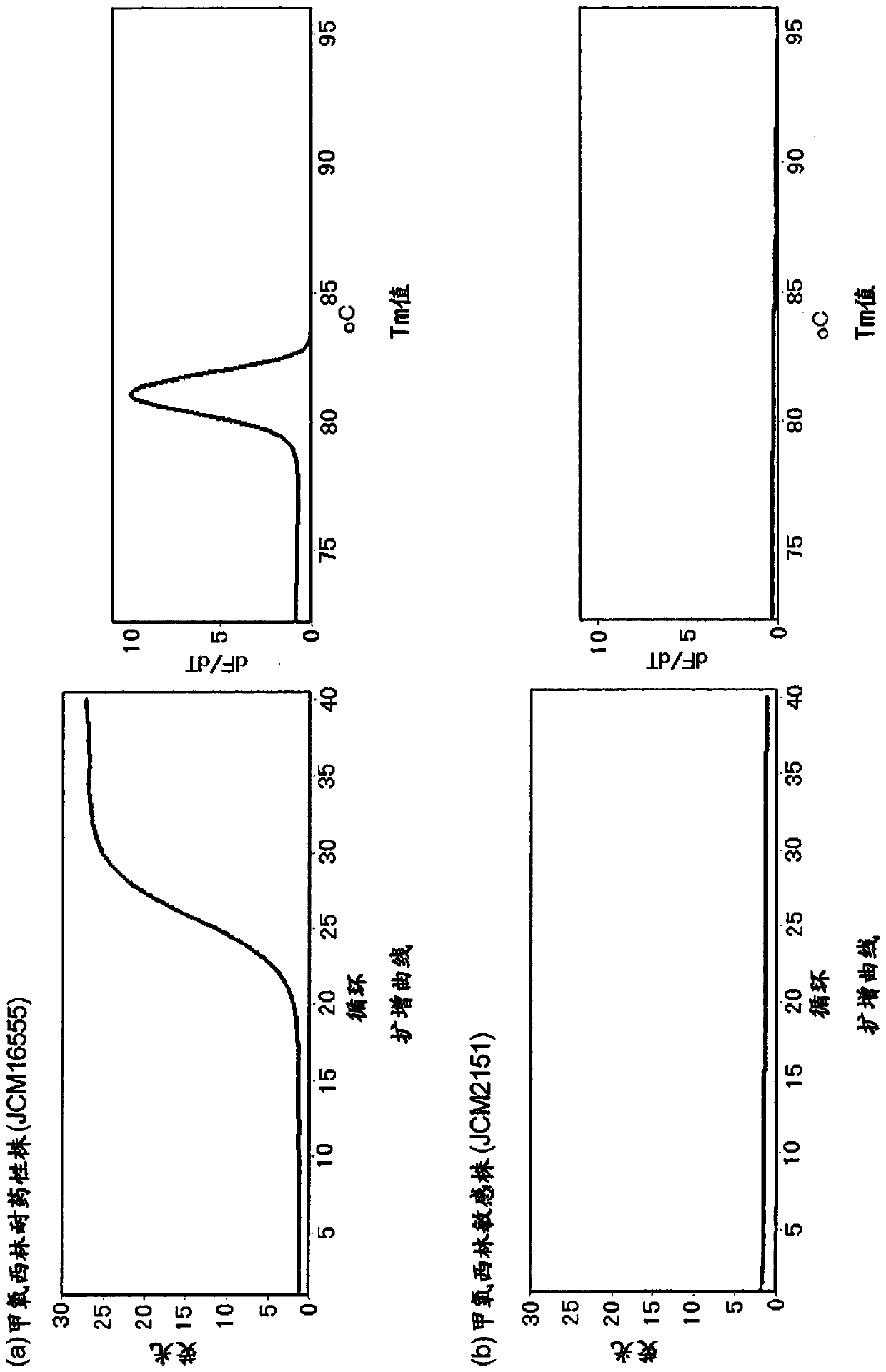
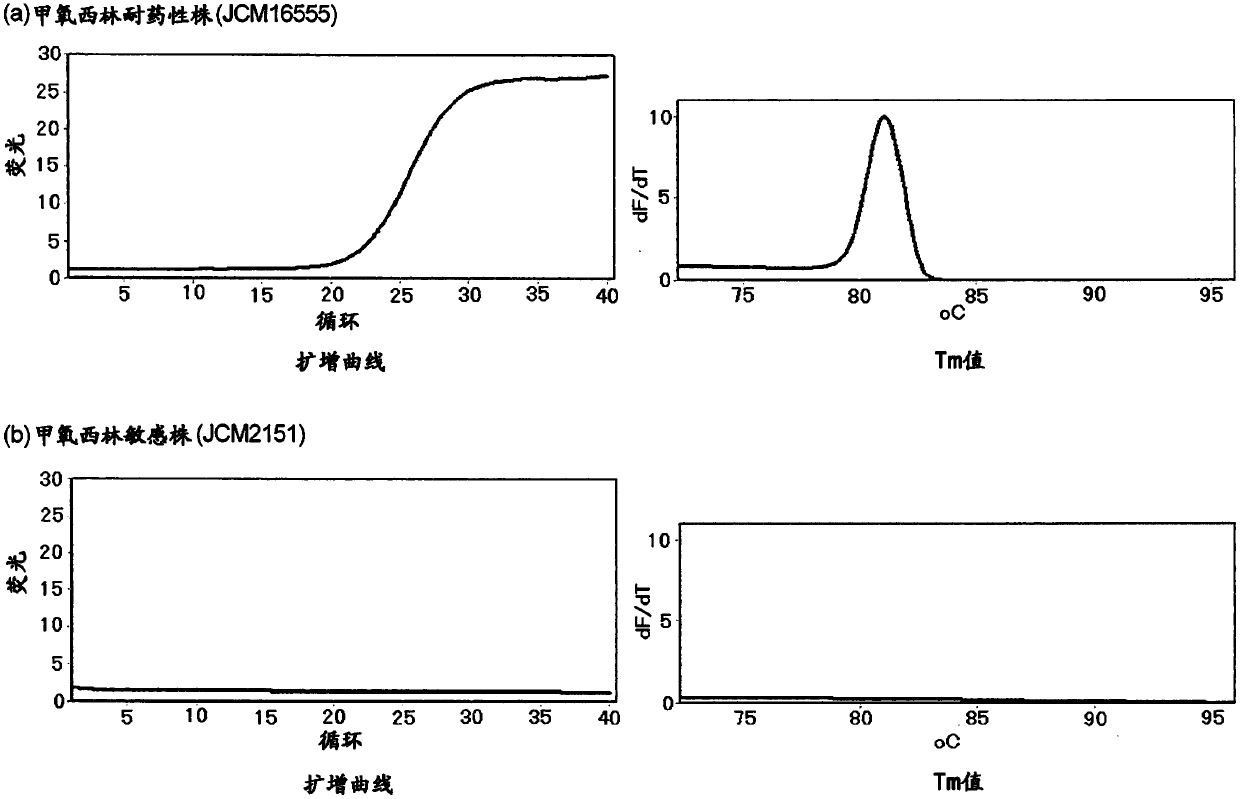
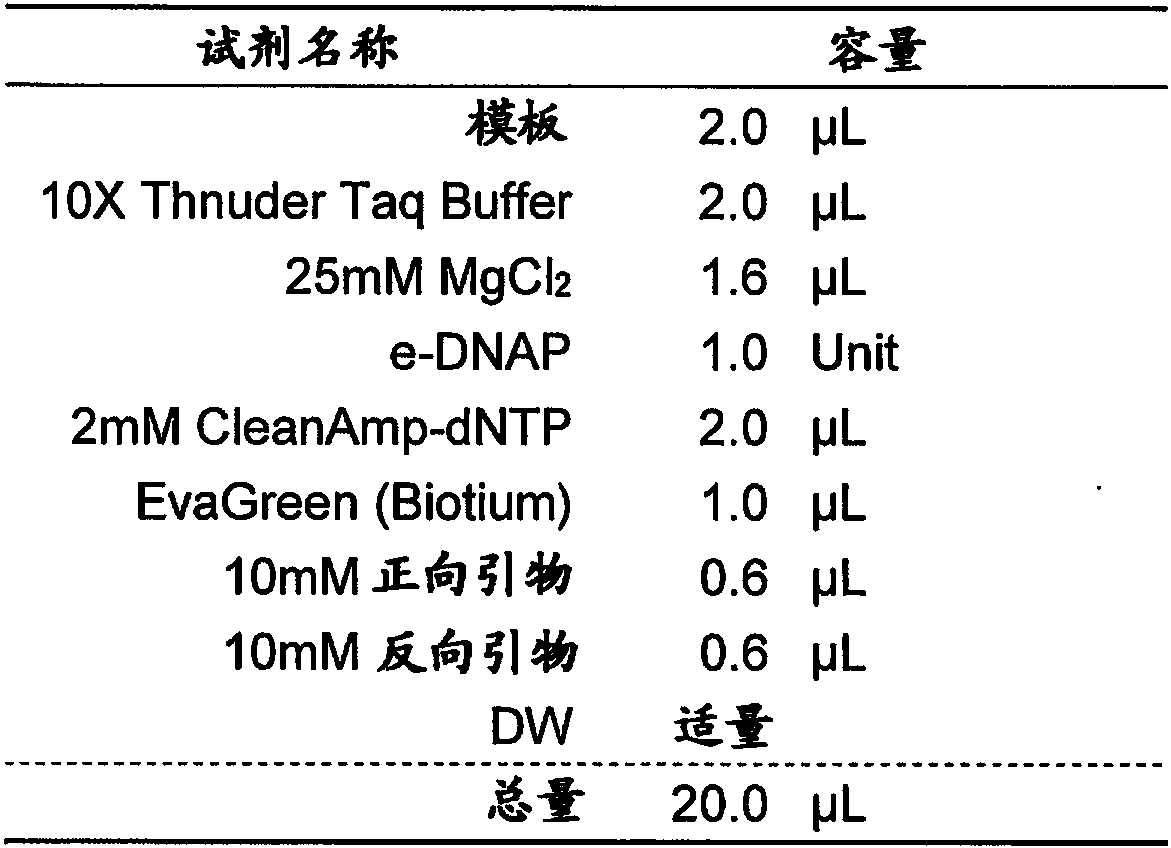
[0165] Genomic DNA extracted from the culture medium of MRSA (RIKEN CORPORATION transfer strain, JCM31453) was used as a template using the High pure PCR template preparation kit (Roche), primers of sequence number 15 and sequence number 16 were used, and the reaction solution shown in Table 1 was used The composition implements PCR to amplify the full length of mecA ORF. Then, a serial dilution of the amplification product of mecA was prepared, and a primer pair for comparison consisting of a forward primer consisting of SEQ ID NO: 13 and a reverse primer consisting of SEQ ID NO: 14 was used, and the reaction solution shown in Table 1 was used. PCR was performed to determine the concentration of the amplified product of mecA whose number of cycles was increased to about 20 cycles, and used as a template. It should be noted that the methicillin-resistant Staphylococcus aureus JCM31453 can be obtained through the Microbial Materials Devel...
Embodiment 2
[0167] (Example 2) secondary screening
[0168] Using 50 μg / ml human genomic DNA solution (invirogen), the concentration of genomic DNA extracted from the culture solution of MRSA (RIKEN CORPORATION transfer strain, JCM31453) using High pure PCR template preparation kit (Roche) was adjusted to increase the number of cycles to about 20 cycles of the solution, used as a template.
[0169] For 88 primer pairs, PCR was performed using the reaction solution composition shown in Table 1, and evaluation was performed. As the real-time PCR device, Rotor-Gene Q MDx 5plex HRM (QIAGEN) was used. The reaction conditions of PCR were: heating at 95° C. for 5 minutes, followed by 40 repetitions of 94° C. for 10 seconds, 65° C. for 10 seconds, and 72° C. for 30 seconds. In terms of evaluation, 57 pairs were screened that were confirmed to be improved compared to the results of the comparative primer pairs with respect to the number of rising cycles, the shape of peaks in HRM, the intensity ...
Embodiment 3
[0170] (Example 3) three screening
[0171]Using 50 μg / ml human genomic DNA solution (invirogen), the concentration of genomic DNA extracted from the culture solution of MRSA (RIKEN CORPORATION transfer strain, JCM31453) using High pure PCR template preparation kit (Roche) was adjusted to 10.4 molecules / reaction solution solution as a template. The 57 primer pairs were evaluated by performing PCR by triple assay using the reaction solution composition shown in Table 1. As the real-time PCR device, Rotor-Gene QMDx 5plex HRM (QIAGEN) was used. The reaction conditions of PCR were: heating at 95° C. for 5 minutes, followed by 40 repetitions of 94° C. for 10 seconds, 65° C. for 10 seconds, and 72° C. for 30 seconds. For evaluation, 9 pairs were screened where amplification was observed in both triple assays, peaks for the height of dF / dT in HRM (lane A), the presence of primer dimers, and the presence or absence of non-specific products were confirmed An improved primer pair com...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com