Microflora specific function gene diversity analysis primer pairs and analysis method
A technology for diversity analysis and microbial community, applied in the field of microbial community-specific functional gene diversity analysis primer pairs and analysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0069] The present invention provides three functional genes and primers thereof for the diversity analysis of specific functional genes of microbial communities, the specific circumstances are as follows:
[0070] A pair of primers for amplifying the nifH gene of a nitrogen-fixing microbial community, the specific sequence of which is:
[0071] Primer F (nifH): AAAGGYGGWATCGGYAARTCCACCAC (SEQ ID NO. 1)
[0072] Primer R (nifH): TTGTTSGCSGCRTACATSGCCATCAT (SEQ ID NO. 2)
[0073] A pair of primers for amplifying the nirS gene of a denitrifying microbial community, the specific sequence of which is:
[0074] Primer F (Cd3a): GTSAACGTSAAGGARACSGG (SEQ ID NO.3)
[0075] Primer R (R3cd): GASTTCGGRTGSGTCTTGA (SEQ ID NO.4), a primer pair for amplifying the nirK gene of denitrifying microbial communities, the specific sequence is:
[0076] Primer F (nirK1aCu): ATCATGGTSCTGCCGCG (SEQ ID NO.5)
[0077] Primer R (nirKR3Cu): GCCTCGATCAGRTTGTGGTT (SEQ ID NO. 6)
[0078] Wherein, in th...
Embodiment 2
[0118] Take a soil sample, extract DNA, and add three kinds of corresponding functional gene primers to amplify the corresponding functional gene. The steps include:
[0119] (1) The first round of amplification:
[0120] The amplification system is:
[0121] KOD Enzyme Amplification System 50μL 10×Buffer KOD 5μL 2mM dNTPs 5μL 25mM MgSO 4
3μL Primer F (10μΜ) 1.5μL Primer R (10μΜ) 1.5μL KOD enzyme 1μL template 1-5μL (ie 100ng) h 2 o
Make up 50μL
[0122] The amplification program is: 94°C, 2min; 98°C, 10s; 58-66°C, 30s; 68°C, 30s; 30 cycles; 68°C, 5min.
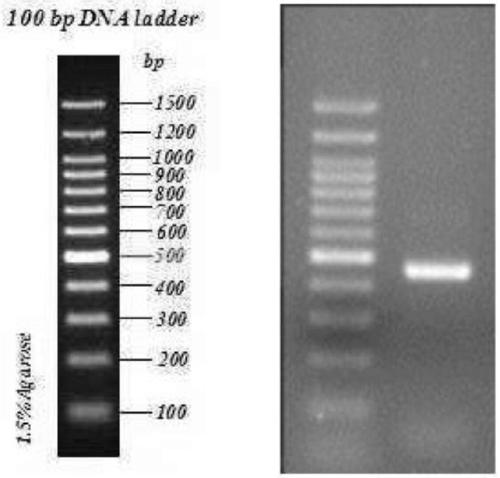
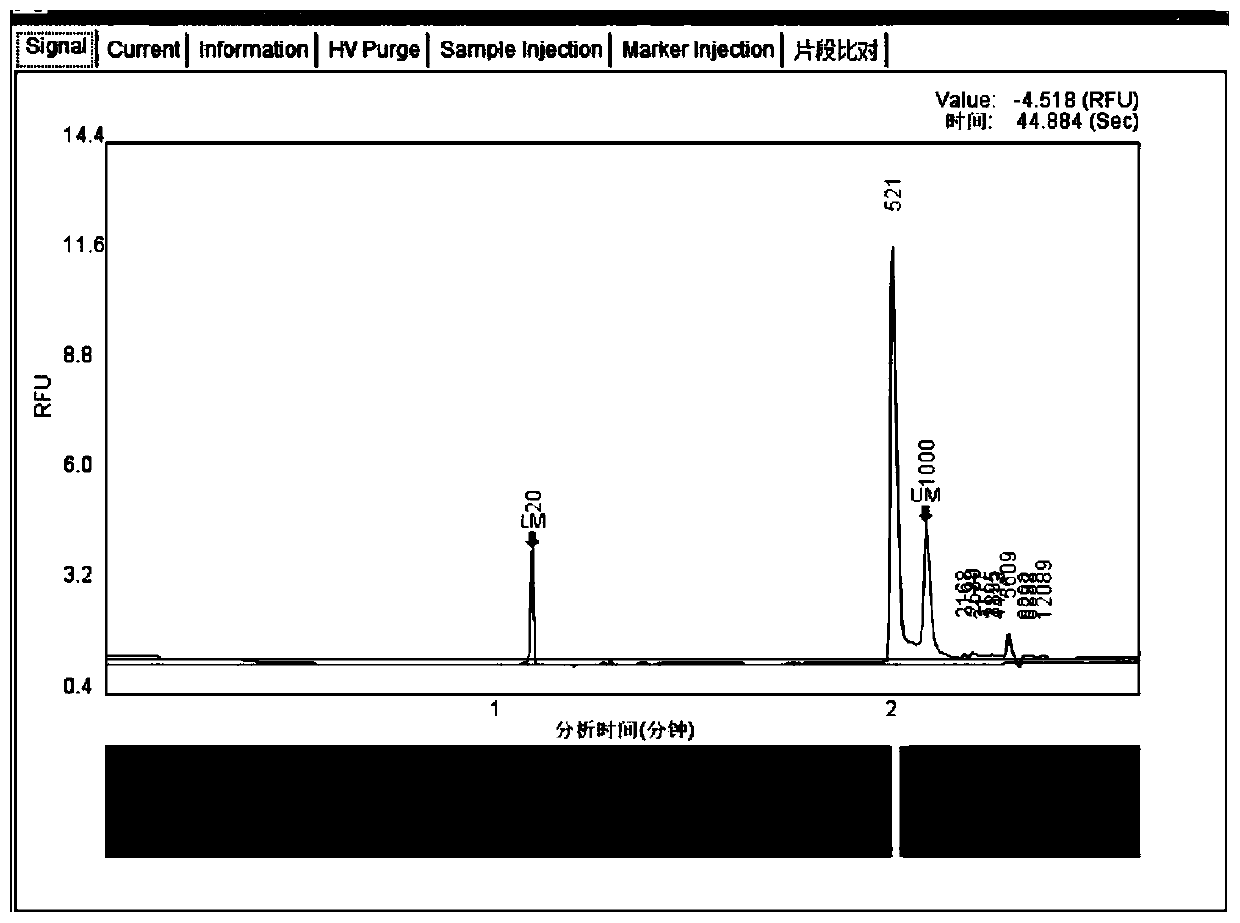
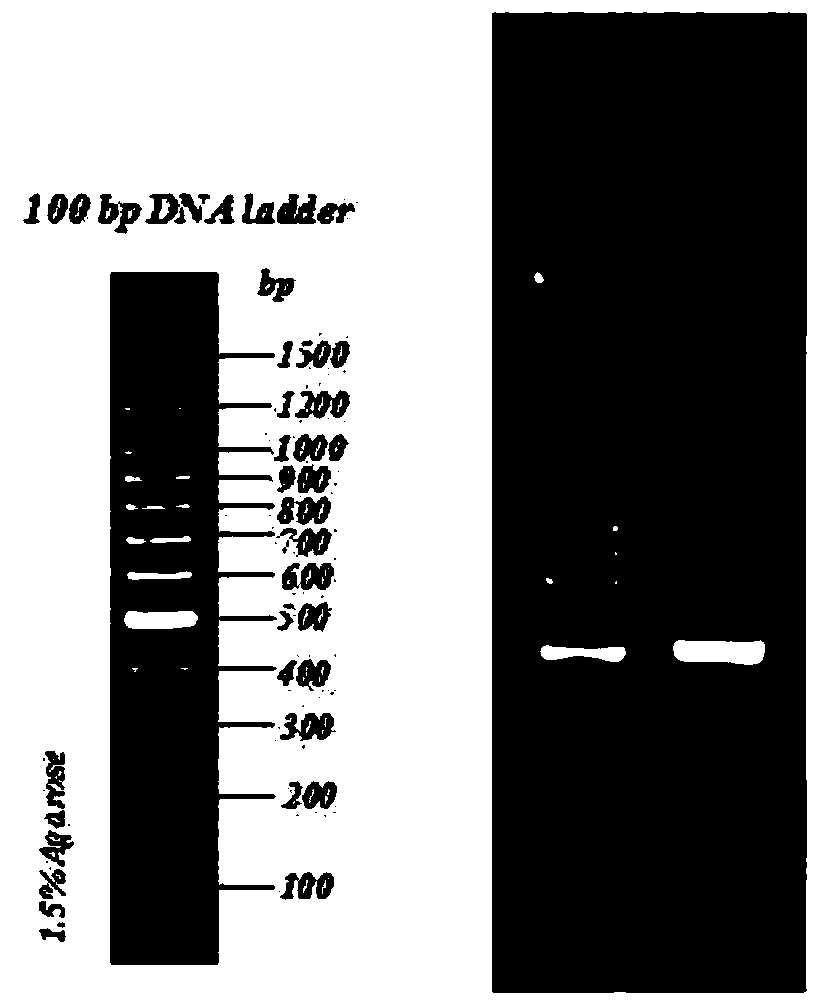
[0123] (2) The first round of amplification product purification includes: using AMPure XP Beads for PCR product purification, after purification, use Qubit3.0 to quantify. Amplification results see figure 1 , image 3 and Figure 5 , PCR amplified bands showed that the specificity of the corresponding functional gene primer amplification was ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com