Primer composition, kit and method for detecting novel coronavirus
A primer composition and coronavirus technology, applied in the field of molecular biology, can solve the problems of complex products, not easy simultaneous analysis of multiple sequences, and complex applications.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
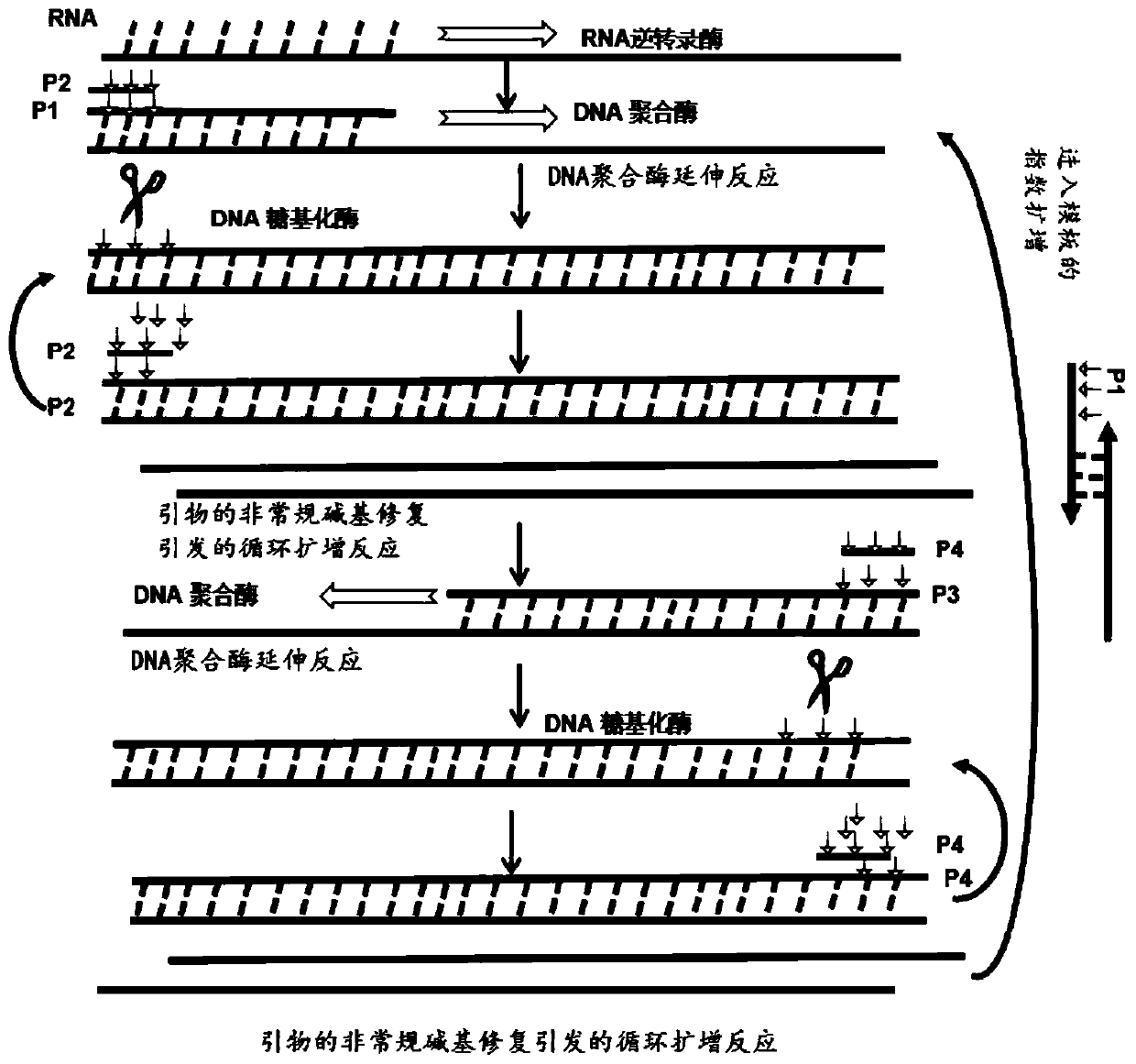
[0063] Example 1 Amplification and detection of N gene and ORF1ab gene of 2019-nCoV The plasmid DNA containing N gene and ORF1ab gene of 2019-nCoV was used as detection target for screening dominant primers.
[0064]Wherein, the conserved sequence of the novel coronavirus N gene is shown in SEQ ID NO:25, and the conserved sequence of the novel coronavirus ORF1ab gene is shown in SEQ ID NO:26.
[0065] SEQ ID NO: 25
[0066] ATTGCCAAAAGGCTTCTACGCAGAAGGGAGCAGAGGCGGCAGTCAAGCCTCTTCTCGTTCCTCATCACGTAGTCGCAACAGTTCAAGAAATTCAACTCCAGGCAGCAGTAGGGGAACTTCTCCTGCTAGAATGGCTGGCAATGGCGGTGATGCTGCTCTTGCTTTGCTGCTGCTTGACAGATTGAACCAGCTTGAGAGCAAAATGTCTGGTAAAGGCCAACAACAACAAGGCCAAACTGTCACTAAGAAATCTGCTGCTGAGGCTTCTAAGAAGCCTCGGCAAAAACGTACTGCCACTAAAGCATACAATGTAACACAAGCTTTCGGCAGACGTGGTCCAGAACAAACCC
[0067] SEQ ID NO: 26
[0068] TGGTGCATCGTGTTGTCTGTACTGCCGTTGCCACATAGATCATCCAAATCCTAAAGGATTTTGTGACTTAAAAGGTAAGTATGTACAAATACCTACAACTTGTGCTAATGACCCTGTGGGTTTTACACTTAAAAACACAGTCTGTACCGTCTGCGGTATGTGGAAAGGTTATGGCTGTA...
Embodiment 2
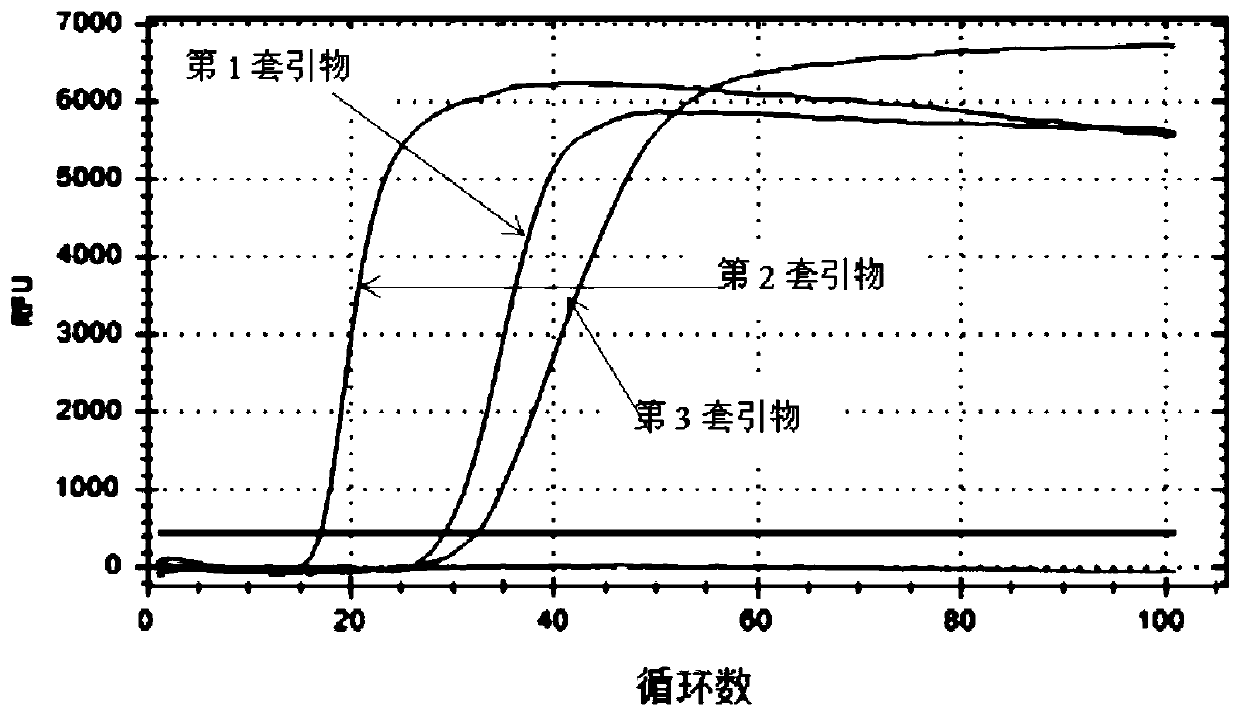
[0085] The reaction efficiency verification of embodiment 2 different primers
[0086] Preparation of an isothermal amplification reaction mixture containing Mg 2+ The concentration is 8mM; K + The concentration is 6mM; NH 4 + The concentration is 10mM; H +The concentration is 20mM; Cl - The concentration is 6mM; SO 4 2- The concentration of Tris-HCl is 10mM; the concentration of Tris-HCl is 20mM; the concentration of Triton X-100 is 0.01g / mL; the concentration of dNTP is 1.4mM; the concentration of thymine DNA glycosylase is 50U / mL; The concentration is 320U / mL; the concentration of MLV reverse transcriptase is 150U / mL; the primers shown in P1 in Table 1 are 0.2 μM, the primers shown in P2 are 0.8 μM, the primers shown in P3 are 0.2 μM, and the primers shown in P4 0.8 μM, and SYBRGreen I is a dye for real-time fluorescence analysis. Using the 3 sets of primers obtained after design, screening and comparison, use the N gene and ORF1ab gene plasmid DNA at a concentratio...
Embodiment 3
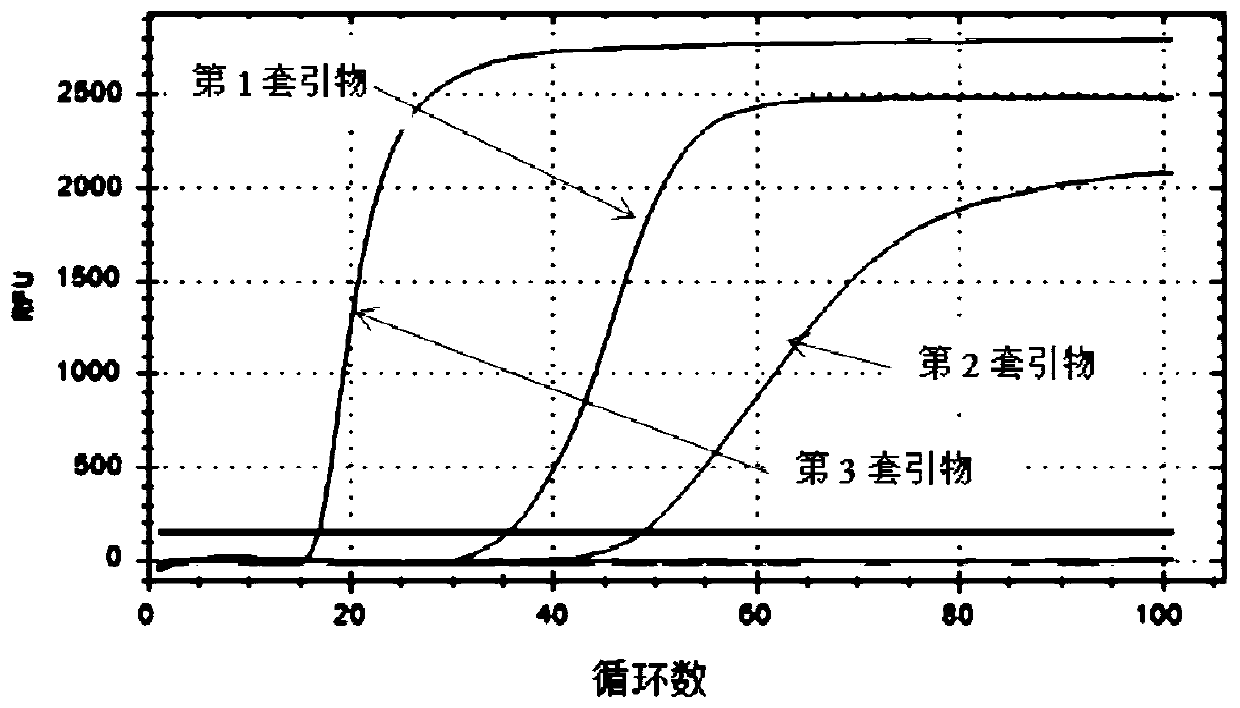
[0087] Example 3 Reaction Sensitivity Verification
[0088] The amplification detection mixture was prepared as described in Example 2, wherein the second set of N gene primers was used to perform amplification and real-time fluorescence detection of the real-time fluorescence curves of different concentrations of N gene plasmid DNA at 63°C and 90 min, and the concentrations were sequentially It is 10 pM, 1 pM, 100 fM, 10 fM, 1 fM, 100 aM, 0 aM. Use a real-time fluorescent quantitative PCR instrument for detection, and read the fluorescence value every 30s. For the real-time fluorescence curve of the specific results, please refer to the attached Figure 4 . The real-time fluorescence curve shows that the concentration of N gene plasmid DNA can be detected as low as 10aM in this example, indicating that the detection method of the present invention has high sensitivity and extremely low detection limit.
[0089] The amplification detection mixture was prepared as described i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com