Method for detecting PCR amplification product
A detection method and a technique for amplifying products, which are applied in biochemical equipment and methods, and the determination/inspection of microorganisms, etc., can solve problems such as the inability to quickly identify the presence or absence of PCR reaction products, achieve low cost, reduce experimental pollution, and simplify experiments program effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
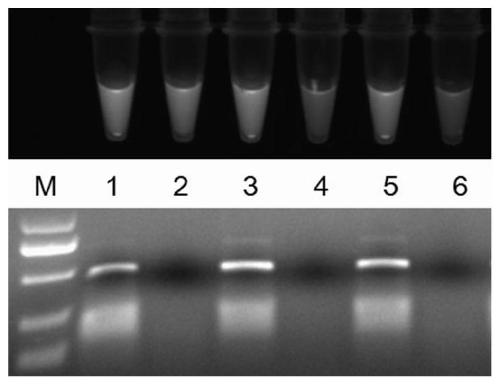
[0025] Example 1 Adding different concentrations of Trion X-100 to the impact of PCR result identification
[0026] Take anticoagulant blood from male pigeons, and use Dzup (blood) Genomic DNA Rapid Extraction Kit (Shanghai Shenggong) to extract DNA according to the instructions. With pigeon DNA as a template, with pigeon CHD gene primers F (5'-AGAACGTGGCAACAGAGTAC-3', the nucleotide sequence is shown in SEQ ID NO: 1) and R (5'-TGAAATGATCCAGTGCTTGT-3', the nucleotide sequence is as shown in Shown in SEQ ID NO:2) carry out PCR amplification. Composition of PCR reaction system (50 μL): 2×Master Mix 25 μL, 10 μM CHD primers F and R 1 μL each, fluorescent dye 4S Red Plus 10000 × stock solution diluted 800 times and then take 2 μL, for comparison with and without Trion X-100 For the impact on the identification of PCR results, samples 1, 3, and 5 were added with 1 μL of pigeon blood DNA template, and samples 2, 4, and 6 were not added as a control, and the volume concentration was...
Embodiment 2
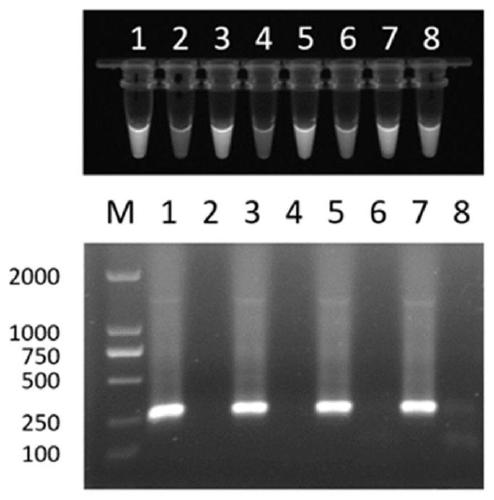
[0027] Embodiment 2 adds Trion X-100 to directly identify the presence or absence of PCR reaction
[0028]The anticoagulant blood of male and female pigeons was collected respectively, and the DNA was extracted with the Dzup (blood) Genomic DNA Rapid Extraction Kit (Shanghai Sangong) according to the instructions. Using pigeon DNA as a template, with female pigeon W chromosome specific primer EE0.6P260 F (5'-TCCTATGCCTACCACCTTCC-3', the nucleotide sequence is shown in SEQ ID NO: 3) and R (5'-ATGATTCTAAAACATTTGTTGTTTG-3', The nucleotide sequence is shown in SEQ ID NO: 4) for PCR amplification. Since male pigeons have no W chromosome, only female pigeons can amplify a 260bp band. Composition of PCR reaction system (50 μL): 2×MasterMix 25 μL, pigeon blood DNA template 1 μL, 10 μM primers EE0.6P260 F and R 1 μL each, fluorescent dye SYBR GREEN I10000 × stock solution diluted 800 times and take 2 μL, the volume concentration is 5% Add 2, 3, 4, and 5 μL of Trion X-100 respectively,...
Embodiment 3
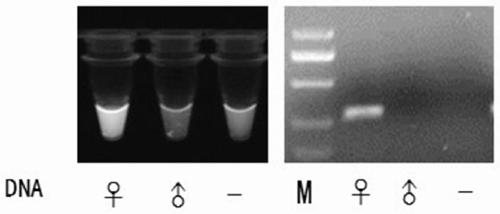
[0029] Embodiment 3 adds NP-40 to directly identify the presence or absence of PCR reaction
[0030] Anticoagulant blood was taken from male and female pigeons respectively, and DNA was extracted with the Dzup (blood) Genomic DNA Rapid Extraction Kit (Shanghai Sangong) according to the instructions. Using pigeon DNA as a template, with female pigeon W chromosome specific primer EE0.6P260 F (5'-TCCTATGCCTACCACCTTCC-3', the nucleotide sequence is shown in SEQ ID NO: 3) and R (5'-ATGATTCTAAAACATTTGTTGTTTG-3', The nucleotide sequence is shown in SEQ ID NO: 4) for PCR amplification. Since male pigeons have no W chromosome, only female pigeons can amplify a 260bp band. PCR reaction system (50 μL) composition: 2×MasterMix 25 μL, pigeon blood DNA template 1 μL, 10 μM primers EE0.6P260 F and R 1 μL each, 4S Red Plus 10000× stock solution diluted 800 times to take 2 μL, 5% NP-40 3 μL, Sterilized deionized water 17 μL. PCR reaction conditions: ①94°C for 3min; ②94°C for 30s; 53°C for 30...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com