Cancer screening method based on protein marker and artificial intelligence
A protein and cancer technology, applied in the field of bioinformatics, can solve the problems of low specificity and sensitivity of tumor markers, can not meet the requirements of clinical early screening, and achieve the effect of improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
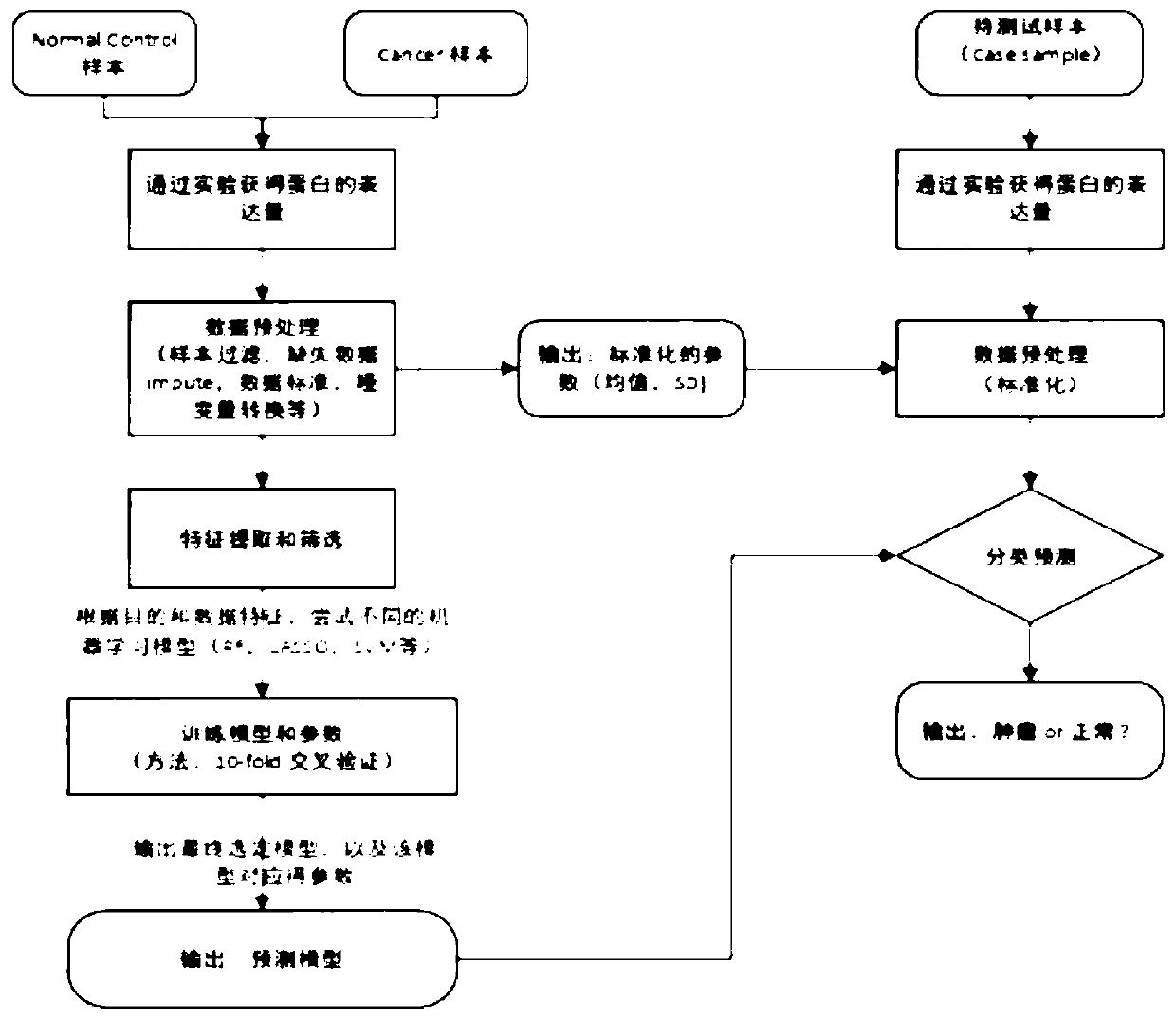
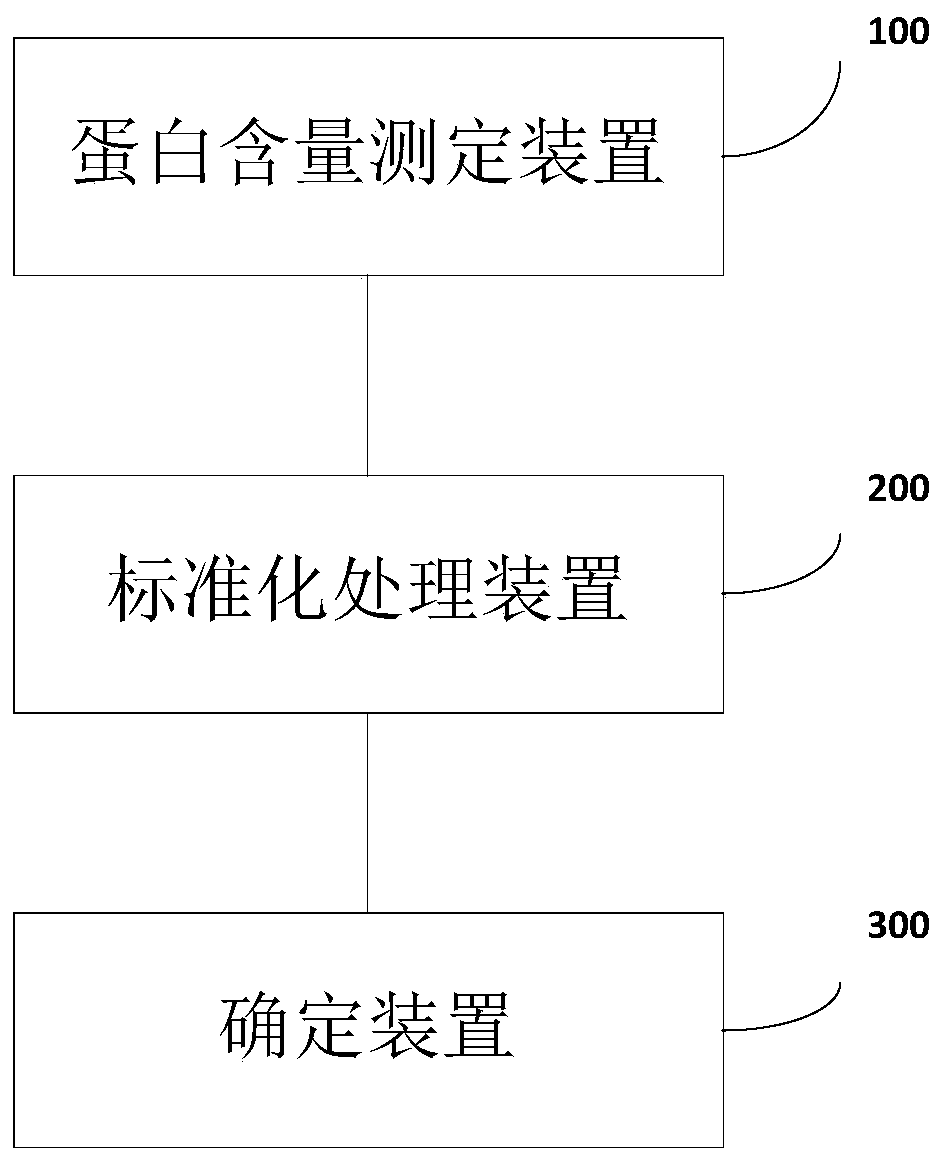
[0066] The common quantitative methods of tumor markers are: 1) Serological level uses radioimmunoassay (RIA); 2) Enzyme-linked immunoassay (ELISA); 3) Automatic immunochemical analysis system to quantify. In the present invention, the newly developed standardized method can be used to offset the impact of different quantitative methods and be used across platforms;
[0067] The samples in this embodiment are all quantified using the fully automatic immunochemical analysis system, and the specific processing process is as follows:
[0068] (1) Sample pretreatment: The blood samples of the subjects or control samples (samples of known type) are each 10ml, and the blood samples are transferred from the blood collection tube to the 10mL centrifuge tube in the biological safety cabinet, and the centrifuge tube cover is marked Sample number and date of sample processing. Centrifuge at 1600g at 4℃ for 10min. Collect the centrifuged supernatant in a new 10mL centrifuge tube. Centrifuge at...
Embodiment 2
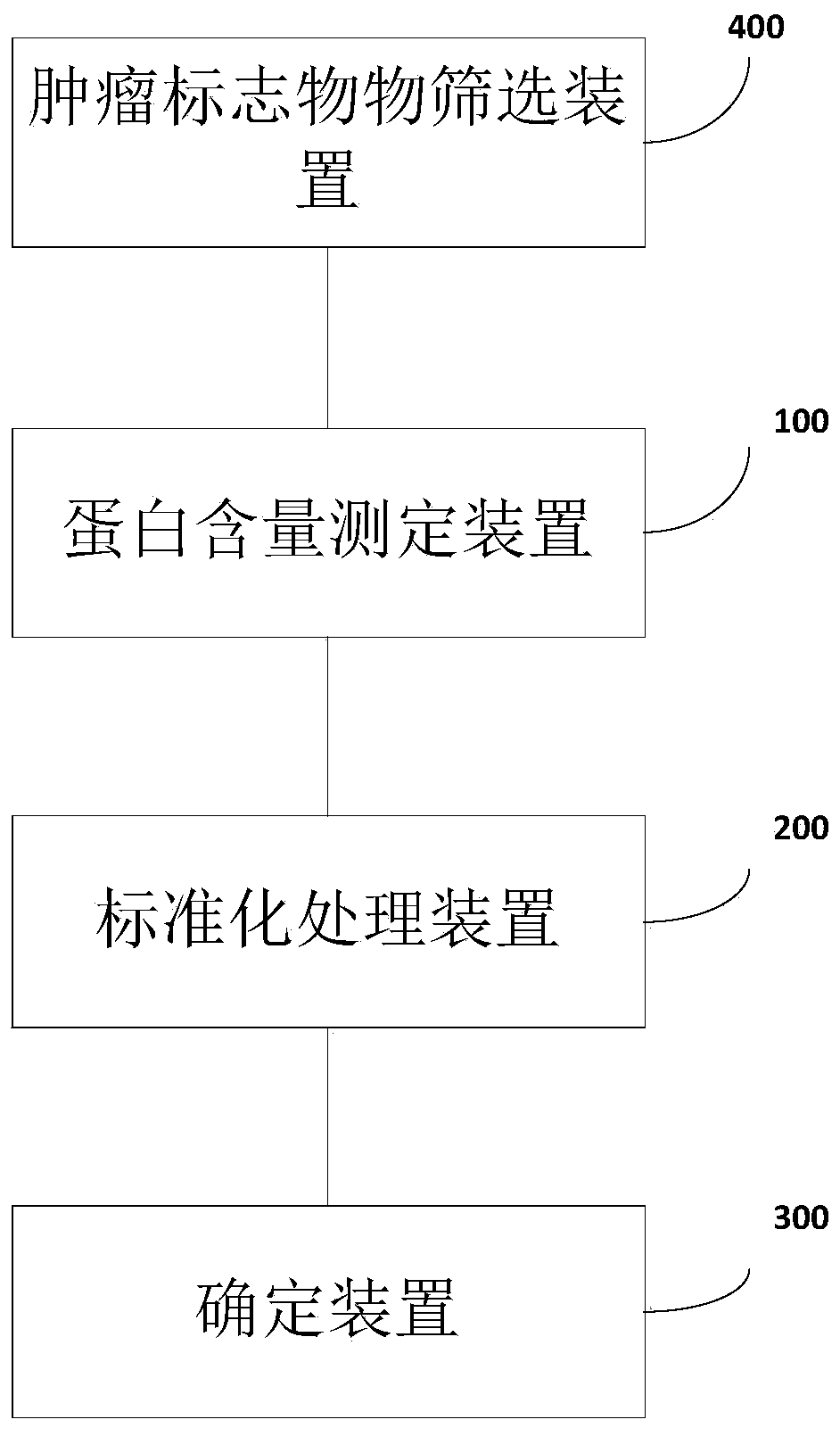
[0072] Marker screening: using the protein tumor marker data in the published literature (Cohen, JD, etal., 2018.), the data contains protein expression data of 1817 samples, of which normal human samples: 812 cases; cancer patients Sample: 1005 cases. The data set includes: AFP, Angiopoietin-2, AXL, CA-125, CA15-3, CA19-9, CD44, CEA, CYFRA 21-1, DKK1, Endoglin, FGF2, Follistatin, NSE, OPG, OPN, PAR, Prolactin, sEGFR, sFas, sHER2 / sEGFR2 / sErbB2, etc. a total of 39 protein marker expression levels.
[0073] (1) Perform t-test comparison between cancer and normal people, and require that the amount of protein table in the cancer group is significantly higher than that in the normal population (excluding 13 protein markers, as shown in Table 1); simultaneously divide different cancers The type (only for a few cancer types with a high incidence of cancer) and the normal sample are subjected to the same hypothesis test. The more cancers with significant differences, the higher the pr...
Embodiment 3
[0086] In order to compare the prediction performance of the method developed by the present invention on early tumor screening, the inventor obtained the detection data of common tumor markers from a third-class hospital as a third-party independent verification test, which was compared with conventional clinical methods. The markers used in this example are clinically routine protein markers: AFP, CEA, CA125, CA153, CA19-9, CYFRA 21-1, etc. The validation set: includes 266 normal physical examination populations and 81 tumor patients with different cancers. Table 3 below lists the quantitative results of tumor markers in 100 samples.
[0087] table 3:
[0088]
[0089]
[0090]
[0091]
[0092] (1) Data filtering and preprocessing: For some samples in Example 1, the detection of all protein markers could not be completed for some reasons or the hospital did some special cancer-related proteins on some samples, and removed the samples. Proportion of protein marker missing> 10% ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com