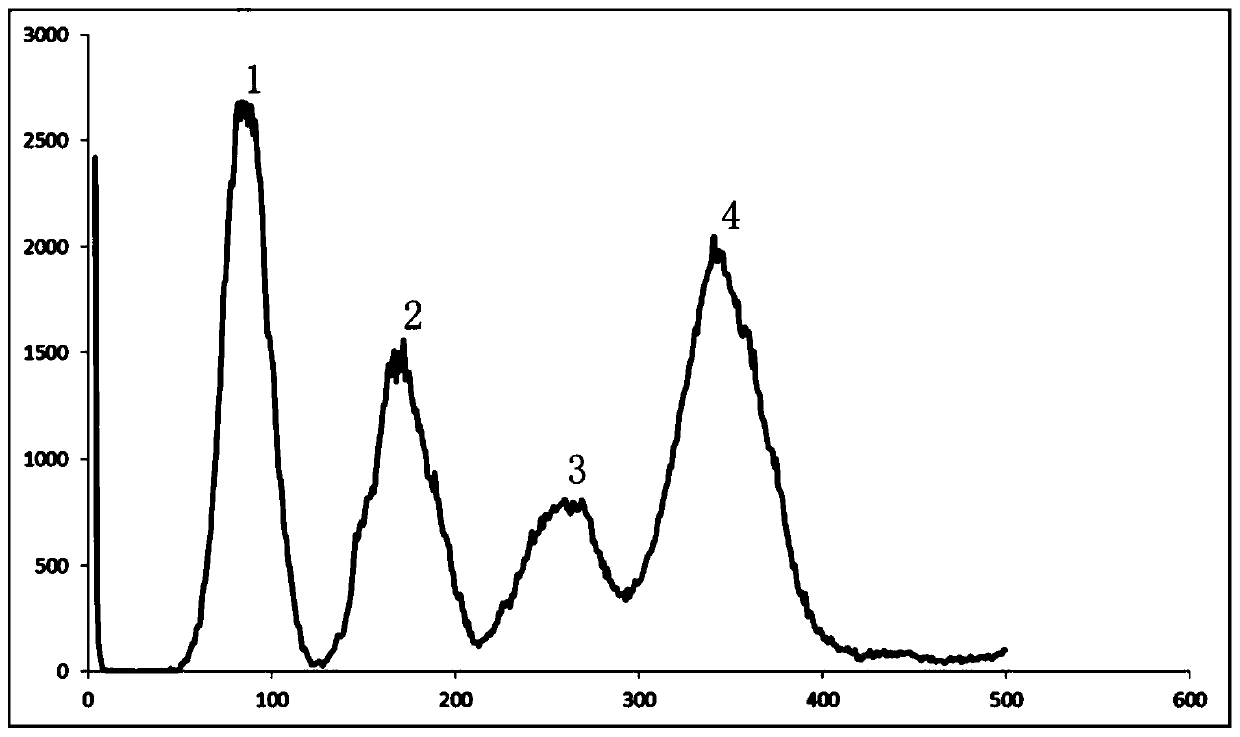
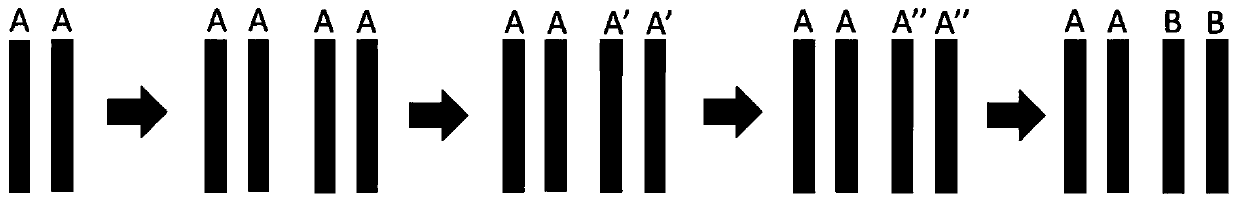
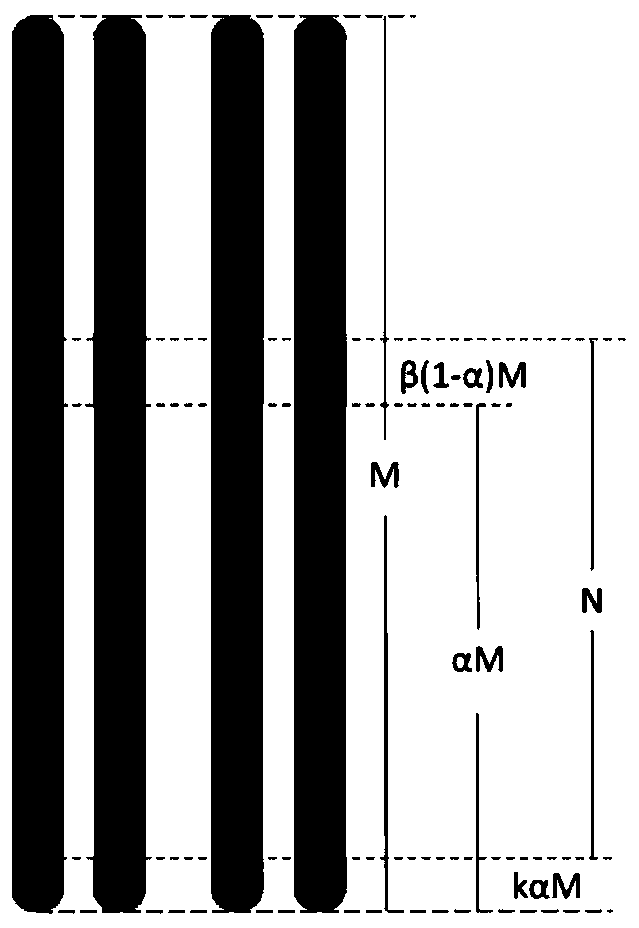
Method for polyploidy genome survey
A genomic and polyploid technology, applied in the field of molecular biology, can solve the problems of low evaluation accuracy of polyploid species, and achieve the effect of improving the evaluation accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047]A method for polyploid genome survey, which specifically includes the following steps:
[0048] Step 1. Genomic DNA extraction and sequencing:
[0049] (1) Cut 10mg of tissue into a 2ml centrifuge tube, cut it with surgical scissors (sterilized with alcohol), and then break it with a homogenizer;
[0050] (2) add 400ul of ACL solution and 20ul of proteinase K;
[0051] (3) Shake and mix for 1 minute, and then place it at 55°C for 3 hours. During this period, take out and mix evenly every half an hour, which is helpful for full cracking. The fully cracked sample should be clear and transparent;
[0052] (4) Take out the sample and shake it evenly when it is lowered to room temperature;
[0053] (5) Add 300ul of Ext solution and 300ul of AB solution to the treated sample in turn, shake vigorously, and then centrifuge at 12000rmp for 5min. The solution will be layered, the upper layer is the blue extraction layer, the lower layer is the transparent water phase, there may...
Embodiment 2
[0078] A method for polyploid genome survey, which specifically includes the following steps:
[0079] Step 1. Genomic DNA extraction and sequencing:
[0080] (1) Cut 7mg of tissue into a 2ml centrifuge tube, cut it with surgical scissors (sterilized with alcohol), and then break it with a homogenizer;
[0081] (2) add 400ul of ACL solution and 20ul of proteinase K;
[0082] (3) Shake and mix for 1 minute, and then place it at 55°C for 3 hours. During this period, take out and mix evenly every half an hour, which is helpful for full cracking. The fully cracked sample should be clear and transparent;
[0083] (4) Take out the sample and shake it evenly when it is lowered to room temperature;
[0084] (5) Add 300ul of Ext solution and 300ul of AB solution to the treated sample in turn, shake vigorously, and then centrifuge at 12000rmp for 5min. The solution will be layered, the upper layer is the blue extraction layer, the lower layer is the transparent water phase, there may...
Embodiment 3
[0109] A method for polyploid genome survey, which specifically includes the following steps:
[0110] Step 1. Genomic DNA extraction and sequencing:
[0111] (1) Cut 10mg of tissue into a 2ml centrifuge tube, cut it with surgical scissors (sterilized with alcohol), and then break it with a homogenizer;
[0112] (2) add 400ul of ACL solution and 20ul of proteinase K;
[0113] (3) Shake and mix for 2 minutes, and then place it at 55°C for 3 hours. During this period, take out and mix evenly every half an hour, which is helpful for full cracking. The fully cracked sample should be clear and transparent;
[0114] (4) Take out the sample and shake it evenly when it is lowered to room temperature;
[0115] (5) Add 300ul of Ext solution and 300ul of AB solution to the treated sample in turn, shake vigorously, and then centrifuge at 12000rmp for 6min. The solution will be layered, the upper layer is the blue extraction layer, the lower layer is the transparent water phase, there m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com