Proteins binding nkg2d, cd16, and c-type lectin-like molecule-1 (cll-1)
A CLL-1, protein technology, applied in the field of multispecific binding proteins
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
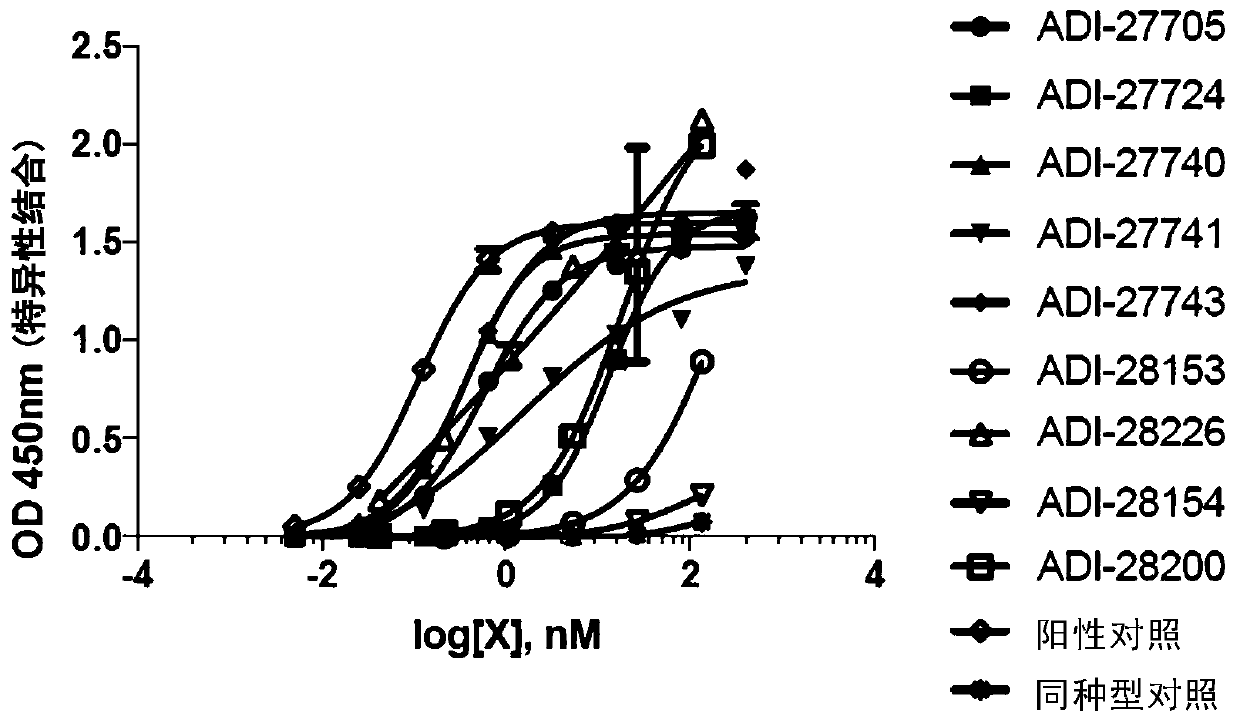
[0249] Example 1 - NKG2D binding domains bind to NKG2D
[0250] NKG2D binding domain binds to purified recombinant NKG2D
[0251] The nucleic acid sequence of human, mouse or cynomolgus NKG2D extracellular domains (ectodomains) is fused with the nucleic acid sequence encoding human IgG1 Fc domain, and introduced into mammalian cells for expression. After purification, the NKG2D-Fc fusion protein was adsorbed to the wells of a microplate. After blocking the wells with bovine serum albumin to prevent non-specific binding, the NKG2D binding domain was titrated and added to the wells pre-adsorbed with NKG2D-Fc fusion protein. Primary antibody binding was detected using a secondary antibody conjugated to horseradish peroxidase and specifically recognizing human kappa light chain to avoid Fc cross-reactivity. The substrate for horseradish peroxidase, 3,3',5,5'-tetramethylbenzidine (TMB), was added to the wells to visualize the binding signal, measured at 450 nM and corrected at 54...
Embodiment 2
[0256] Example 2 - NKG2D Binding Domains Block Natural Ligand Binding to NKG2D
[0257] Competition with ULBP-6
[0258] The recombinant human NKG2D-Fc protein was adsorbed to the wells of a microplate, and the wells were blocked with bovine serum albumin to reduce non-specific binding. A saturating concentration of ULBP-6-His-biotin was added to the wells followed by NKG2D binding domain clones. After 2 hours of incubation, the wells were washed and ULBP-6-His-biotin still bound to NKG2D-Fc-coated wells was detected by streptavidin-coupled horseradish peroxidase and TMB substrate . Absorbance was measured at 450 nM and corrected at 540 nM. The specific binding of the NKG2D binding domain to the NKG2D-Fc protein was calculated from the percentage of ULBP-6-His-biotin blocked from binding to the NKG2D-Fc protein in the wells after background subtraction. Positive control antibodies (comprising heavy and light chain variable domains selected from SEQ ID NO: 101-104) and vari...
Embodiment 3
[0265] Example 3 - NKG2D binding domain cloning activates NKG2D
[0266] The nucleic acid sequences of human and mouse NKG2D were fused to the nucleic acid sequence encoding the CD3ζ signaling domain to obtain chimeric antigen receptor (CAR) constructs. The NKG2D-CAR construct was then cloned into a retroviral vector using Gibson assembly and transfected into expi293 cells for retroviral production. EL4 cells were infected with NKG2D-CAR-containing virus together with 8 μg / mL polybrene. 24 hours after infection, the expression level of NKG2D-CAR in the EL4 cells was analyzed by flow cytometry, and clones expressing high levels of NKG2D-CAR on the cell surface were selected.
[0267] To determine whether NKG2D-binding domains activate NKG2D, they were adsorbed to wells of microplates and NKG2D-CAR EL4 cells were incubated on antibody fragment-coated wells in the presence of brefeldin-A and monensin Incubate for 4 hours. Intracellular TNF-[alpha] production, an indicator of N...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com