Bombyx mori actin 6A gene and application in detection of methylation level of bombyx mori actin 6A gene
An actin and methylation technology, applied in application, genetic engineering, plant genetic improvement, etc., can solve the problems of difficult control, strong virus infectivity, frequent outbreaks, etc., and achieve the effect of high stability and strong specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1D
[0023] Example 1DNA Extraction
[0024] Use Ezup Column Animal Genomic DNA Purification Kit to extract DNA, the specific steps are as follows:
[0025] (1) The experimental group was BmN cells added with Bombyx mori nuclear polyhedrosis virus, and the control group was normal BmN cells. Add 200 μl Buffer PBS and 20 μl Proteinase K to each well of cells, shake the culture flask gently, collect the cells, and transfer them to a new Add 200μl Buffer CL to the centrifuge tube, shake and mix, and bathe in 56℃ water for 10min;
[0026] (2) Add 200μl Buffer CL and mix thoroughly by inversion;
[0027] (3) Add 200 μl of absolute ethanol, fully invert and mix;
[0028] (4) Put the adsorption column into the collection tube, add all the solution and translucent fibrous suspension into the adsorption column with a pipette, let it stand for 2 minutes, then centrifuge at 10,000 rpm for 1 minute at room temperature, and pour out the waste liquid in the collection tube;
[0029] (5) Put t...
Embodiment 2
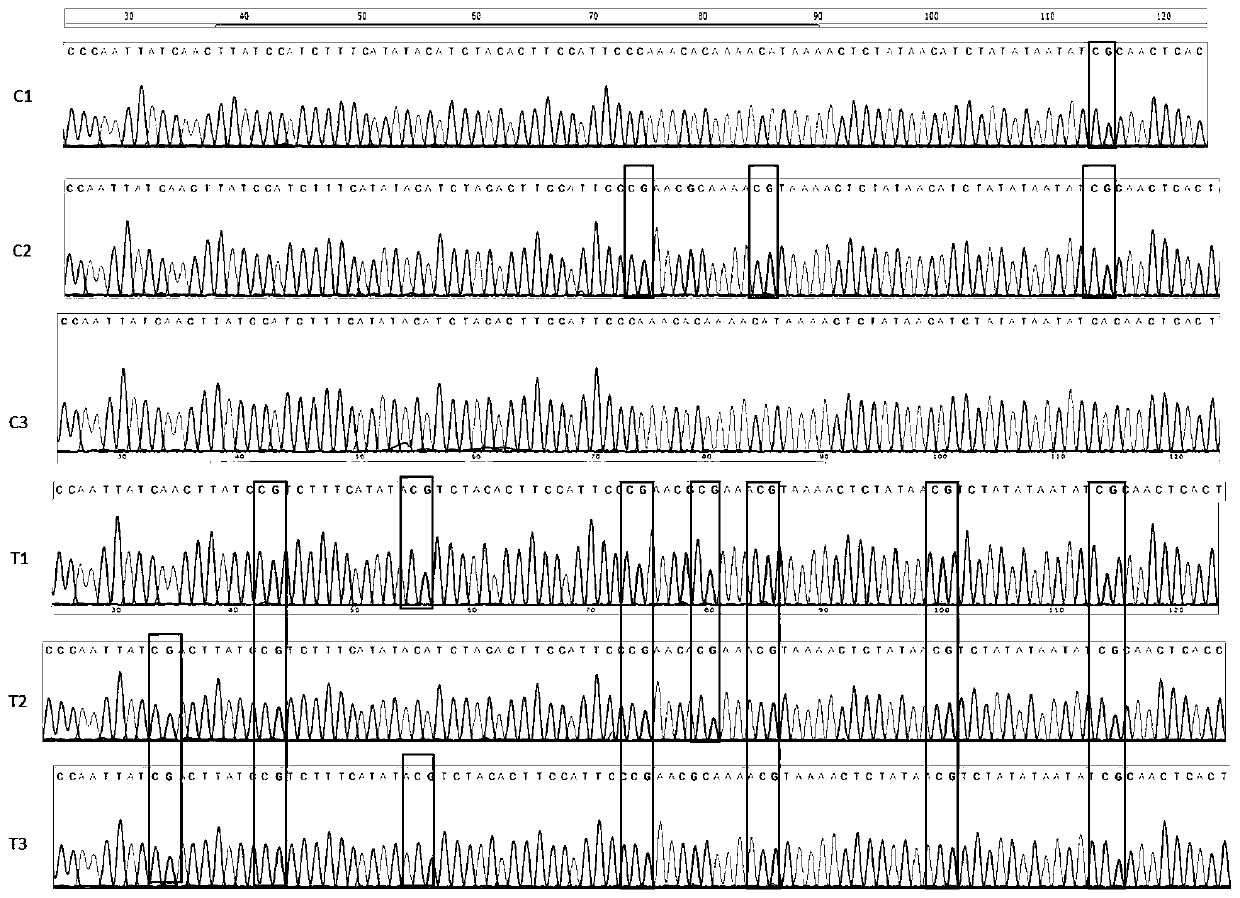
[0033] Example 2 DNA bisulfite treatment
[0034] Use the bisulfite treatment reagent provided in the kit to perform bisulfite modification on the DNA extracted in step 1. The specific steps are as follows:
[0035] (1) Add 130μL of CT Conversion Reagent and 20ul of DNA sample to the single PCR tube (if the sample is less than 20ul, fill it up with water);
[0036] (2) Place the above sample in a water bath on a water bath (98°C for 10min; 64°C for 2.5h);
[0037] (3) Add 600μL of M-Binding Buffer to the Zymo-SpinTMIC Column and put it into the collection tube;
[0038] (4) Add the product after PCR in step 2 to the Zymo-SpinTMIC Column containing M-Binding Buffer, close the lid and mix by inverting;
[0039] (5) Centrifuge at 12000rpm for 30s, discard the liquid in the collection tube;
[0040] (6) Add 100μL of M-Wash Buffer and centrifuge at 12000rpm for 30s;
[0041] (7) Add 200 μL of M-Desulphonation Buffer, let stand at room temperature (20°C-30°C) for 15-20min, and c...
Embodiment 3
[0044] Embodiment 3PCR amplification reaction
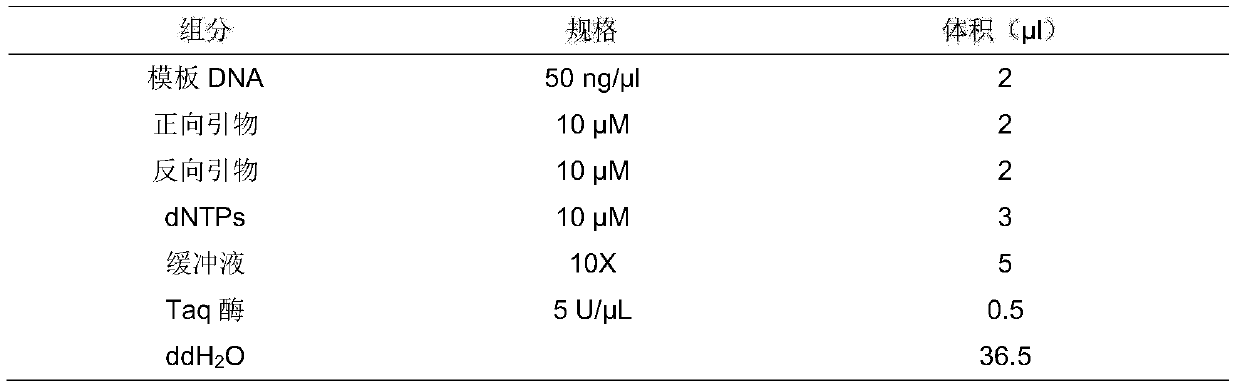
[0045] The amplification reagent provided in the kit was used to carry out PCR amplification using the bisulfite-modified DNA in step 2 as a template. The amplification system is shown in Table 1.
[0046] Table 1 PCR amplification system
[0047]
[0048] The PCR reaction conditions are shown in Table 2.
[0049] Table 2 PCR reaction conditions
[0050]
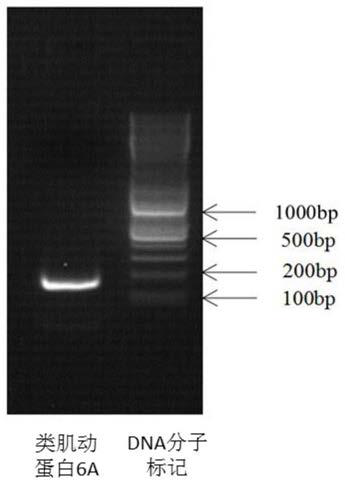
[0051] After the PCR reaction was completed, the agarose gel electrophoresis was detected, and the results showed as follows: figure 1 , indicating that the primers of the present invention can amplify specific target fragments.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com