Primer group for detecting pathogenic gene mutation of pheochromocytoma and paraganglioma and application method of primer group
A technology for pheochromocytoma and paraganglioma, which is applied in the field of primer sets for detecting pathogenic gene mutations in pheochromocytoma and paraganglioma, and can solve the problem of low throughput of pathogenic gene mutation detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0087] Design and synthesize primer sets
[0088] Step 1.1: Design specific upstream and downstream primers based on exons of pheochromocytoma and paraganglioma pathogenic mutation sites in SDHB, SDHC, SDHD, VHL, RET, TMEM127, and MAX genes.
[0089] For the design of primers, Primer Quest and Primer Premier 5.0 were used to design primers and analyze dimers and stem-loop mismatches. Primers were designed at both ends containing mutation sites, and the annealing temperatures of the 20 pairs of primers were basically consistent.
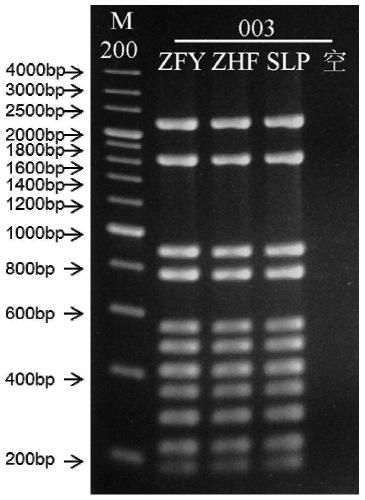
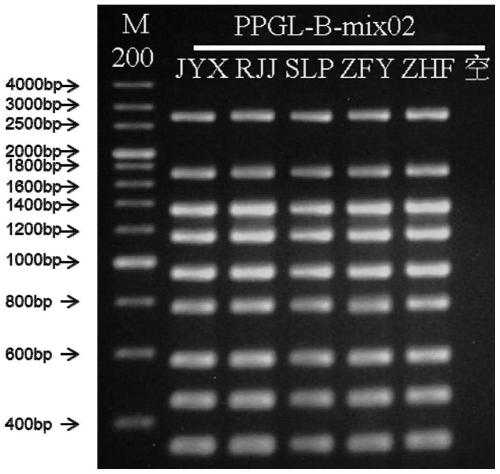
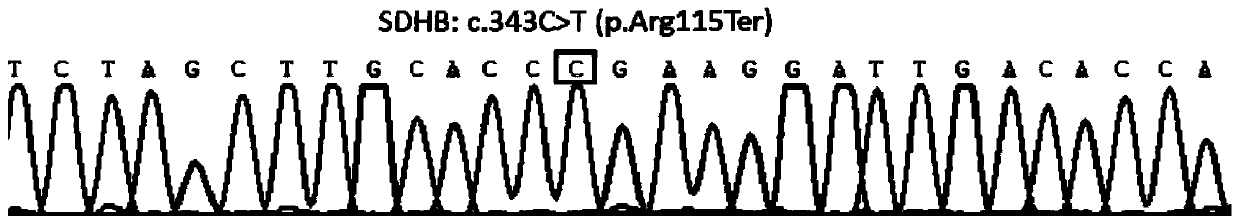
[0090] The primer set provided in this example covers the pathogenic mutation sites of pheochromocytoma and paraganglioma in SDHB, SDHC, SDHD, VHL, RET, TMEM127, and MAX genes. Since small sequence changes will lead to a significant decrease in primer amplification efficiency and poor specificity, multiple PCR primer sets were designed for different sites / exons, and after pre-experimental screening, the length and position of the product fragments wer...
Embodiment 2
[0102] Genomic DNA extraction from samples to be tested
[0103] Step 2.1: Collect oral exfoliated cells or fresh peripheral blood samples with oral swabs.
[0104] In this embodiment, the sample to be tested is a buccal swab sample or a fresh human peripheral blood sample.
[0105] Step 2.2: Use Tiangen Oral Swab Genomic DNA Extraction Kit (DP322), or, use Blood / Cell / Tissue Genomic DNA Extraction Kit (DP304) to extract genomic DNA from the sample, and use NP80-touch (Germany IMPLEN ) to determine the concentration and purity of DNA, and to preserve genomic DNA.
Embodiment 3
[0107] Prepare PCR reaction system
[0108] Step 3.1: Using the genomic DNA obtained in step 2.2 as an amplification template, and using the primer set synthesized in step 1.2, prepare a multiplex PCR reaction system.
[0109] In this example, the DNA polymerase and buffer in the KOD FX enzyme system (article number: KFX-101) of TOYOBO Co., Ltd. were used as the basic raw materials. concentration, buffer concentration, and enzyme dosage to prepare a multiplex PCR amplification system. The specific composition of this reaction system is shown in Table 4 below.
[0110] Table 4
[0111] Reagent components volume 5×PCR buffer for FX 26μl 2mM dNTPs 5μl Primer Mix (1 μM for each primer) 5μl KOD FX (1U / μl) 1μl amplified template 1μl Ultra-pure water 12μl
[0112] The primers were mixed equimolarly, and the total primer concentration was 10 micromolar; the amount of DNA template could be adjusted, and 30 ng of genomic DNA co...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com