Combination of primers and CRISPR sequence for detecting novel coronavirus gene, and application thereof
A primer sequence and gene technology, applied in the field of gene detection, can solve problems such as inconvenient operation and use, and achieve the effects of reducing the probability of missed detection, high sensitivity and easy operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
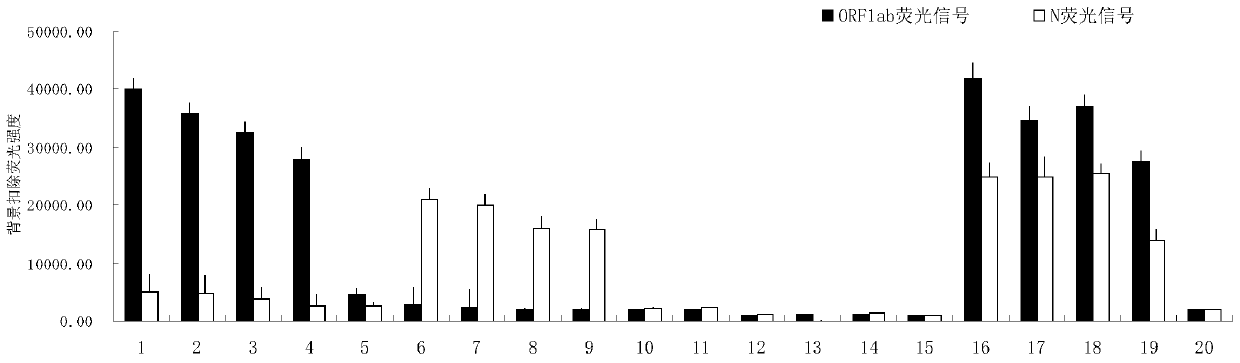
Image
Examples
Embodiment 1
[0018] Embodiment 1 synthetic RPA primer
[0019] RPA amplification primers were synthesized according to the sequence fragments of the selected ORF1ab and N genes, and the primer sequences are shown in Table 1 and Table 2 below. RPA primers were dissolved in RNase-free purified water to form a stock solution (24 μM), and stored at -20°C after aliquoting.
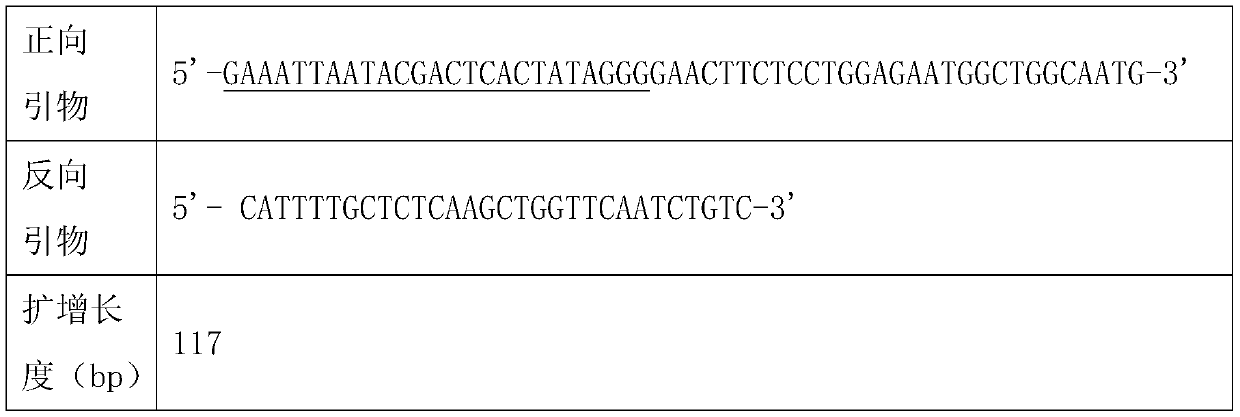
[0020] Table 1: RPA primer sequences for amplifying the ORF1ab gene fragment
[0021]
[0022] Table 2: RPA primer sequences for amplifying N gene fragments
[0023]
[0024]
Embodiment 2
[0025] Embodiment 2, synthetic CrRNA
[0026] Suzhou Synbio was commissioned to synthesize CrRNA, and the sequence information of CrRNAs is shown in Table 3. The synthesized CrRNA was dissolved into 10X stock solution (225nM) with RNase-free purified water, and stored at -20°C after aliquoting.
[0027] Table 3: Synthetic CrRNAs sequences
[0028]
Embodiment 3
[0029] Embodiment 3, the synthesis of reporter probe
[0030] The ssRNA reporter probe used by the Cas13 enzyme of the present invention: entrust Guangzhou Bolaisi to synthesize two ssRNA reporter probes. See Table 4 for sequence specific information. The synthesized ssRNA reporter probes were dissolved in RNase-free purified water to form a stock solution (1.25 μM), and stored at -20°C after aliquoting.
[0031] Table 4: Specific sequence information of reporter probes used by Cas13 protease
[0032]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com