Carbon source absorption expression system, recombinant bacteria and application
A technology of absorption system and recombinant bacteria, applied in the field of genetic engineering, can solve the problems of affecting the yield and fermentation titer of the target product, affecting the industrial application, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1. Construction of carbon source absorption system.
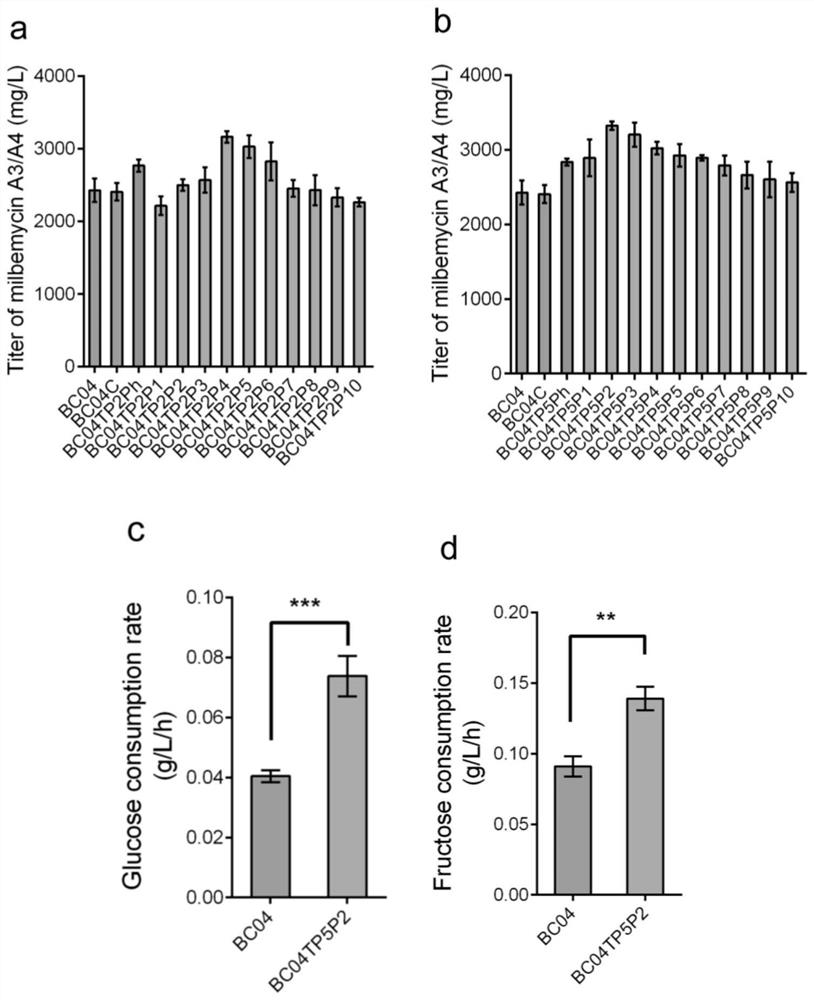
[0027] The carbon source absorption system described in this example is composed of a promoter and a gene for absorbing carbon source connected by a restriction endonuclease, the promoter is promoter P2 or promoter P4, and the gene for absorbing carbon source is Gene TP2 or TP5.
[0028] Using the genome of Streptomyces icebergi BC04 as a template, primers P2-F / P2-R were used to amplify the promoter P2(P sbi_07857 ) sequence, as shown in SEQ ID No.3; Utilize primer P4-F / P4-R to amplify promoter P4 (P sbi_06929 ) sequence, as shown in SEQ ID No.4.
[0029] Using the genome of Streptomyces iceberg BC04 as a template, using primers TP2-F / TP2-R to amplify the gene TP2 that absorbs carbon sources, the TP2 gene contains 3 gene fragments, and the registration of the 3 gene fragments on GenBank The numbers are GeneID: 11614576 (encoding transmembrane protein), 11614577 (encoding transmembrane protein) and 11614578...
Embodiment 2
[0031] Example 2. Construction of a recombinant vector containing the carbon source uptake system described in Example 1.
[0032] Using the genome of Streptomyces icebergi BC04 as a template, primers P1-F / P1-R were used to amplify the promoter P1(P sbi_05102 ) sequence, the P1 promoter and the TP2 gene obtained in Example 1 and EcoRI, XbaI digested linearized pSET152 plasmid backbone using a multi-fragment one-step cloning kit (purchased from Nanjing Nuoweizan Biotechnology Co., Ltd.) through Gibson Assembling method, the plasmid P1-TP2 of the promoter P1 controlling the sugar absorption system was obtained, and the plasmid P1-TP5 of the promoter P1 controlling the sugar absorption system was obtained by referring to the same method.
[0033] The promoter P2 and the plasmid P1-TP2 were respectively digested by NotI and SpeI and ligated to obtain the recombinant vector P2-TP2 of the sugar absorption system controlled by the promoter P2.
[0034] The promoter P4 and the plasmi...
Embodiment 3
[0037] Example 3. Construction of recombinant bacteria that synthesize Streptomyces secondary metabolites.
[0038] The recombinant vectors P2-TP2, P4-TP2, P2-TP5, and P4-TP5 prepared in Example 2 were introduced into Streptomyces glaciferi BC04 by conjugative transfer, respectively, to obtain recombinant strains for the synthesis of milbemycin BC04TP2P2 and BC04TP2P4 and BC04TP5P2 and BC04TP5P4.
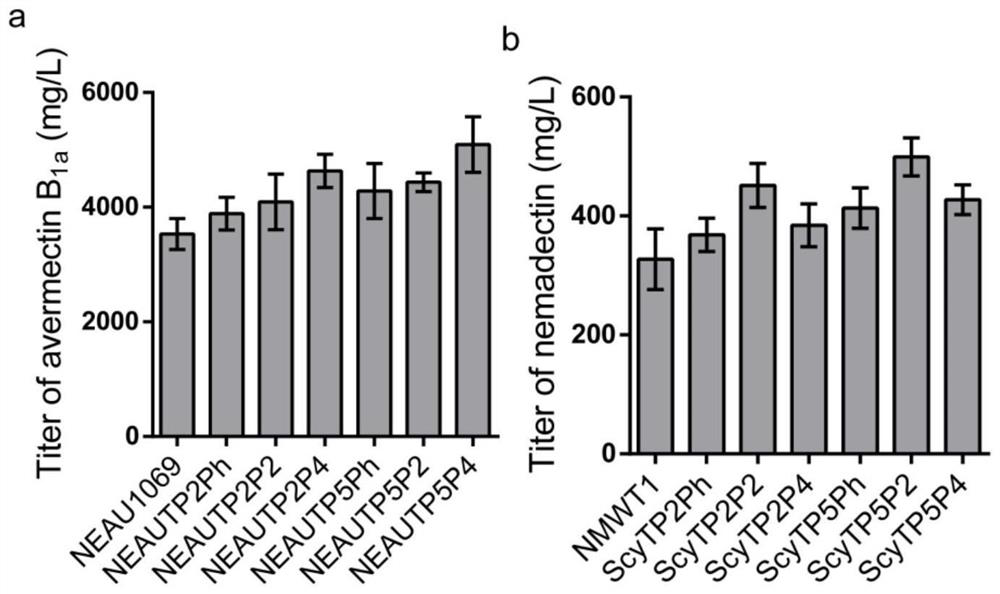
[0039] The recombinant vectors P2-TP2 prepared in Example 2, P4-TP2, P2-TP5 and P4-TP5 were introduced into Streptomyces avermitilis NEAU1069 by conjugative transfer respectively to obtain recombinant strains NEAUTP2P2 for synthesizing abamectin, respectively. NEAUTP2P4, and NEAUTP5P2 and NEAUTP5P4.
[0040]The recombinant vectors P2-TP2, P4-TP2, P2-TP5 and P4-TP5 prepared in Example 2 were introduced into Streptomyces coeruleus NMWT1 by conjugative transfer, respectively, to obtain recombinant strains ScyTP2P2 for the synthesis of nimoctine , ScyTP2P4, ScyTP5P2 and ScyTP5P4.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com