Pyruvate response biosensor as well as construction method and application thereof
A biosensor, pyruvate technology, applied in the field of metabolic engineering, can solve the problems of dynamic control of the metabolic network of difficult strains, difficult to achieve fine control, etc., and achieve the effect of avoiding harmful effects and good specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Embodiment 1: Construction of bacterial strain 168-PR
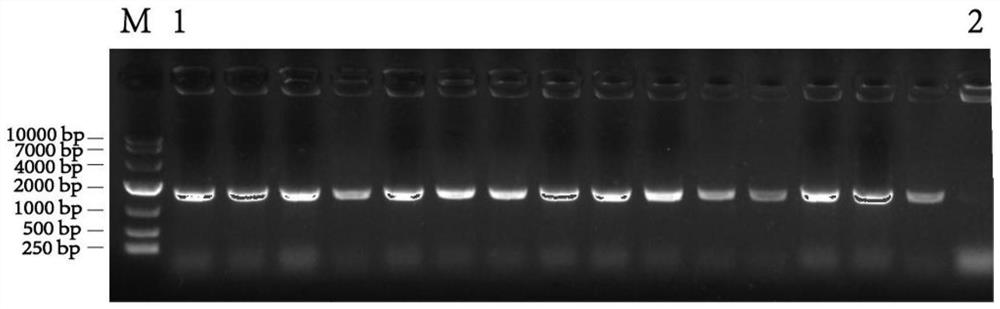
[0043] (1) Using the Escherichia coli K12 genome as a template, P43-pdhR-F and P43-pdhR-R as primers, amplify the pdhR fragment; using the Bacillus subtilis 168 genome as a template, pdhR-FF and pdhR-FR as primers, Amplify the upstream homology arm of pdhR; use pdhR-RF and pdhR-RR as primers to amplify the downstream homology arm of pdhR; artificially synthesize the lox71-spc-lox66-P43 cassette containing the spectinomycin gene and the P43 promoter ( The sequence is shown in SEQ ID NO.12); the pdhR fragment, the pdhR upstream homology arm, the pdhR downstream homology arm and the lox71-spc-lox66-P43 box were fused using fusion PCR technology to obtain the fusion gene fragment pdhRF-lox71-spc -lox66-P43-pdhR-pdhRR (refer to the Takara Primer STARMax DNA Polymeraser manual for the experimental method).
[0044] (2) Transform the fusion fragment obtained in step (1) into Bacillus subtilis 168 by electric shock, the e...
Embodiment 2
[0045] Example 2: Construction of pyruvate biosensor
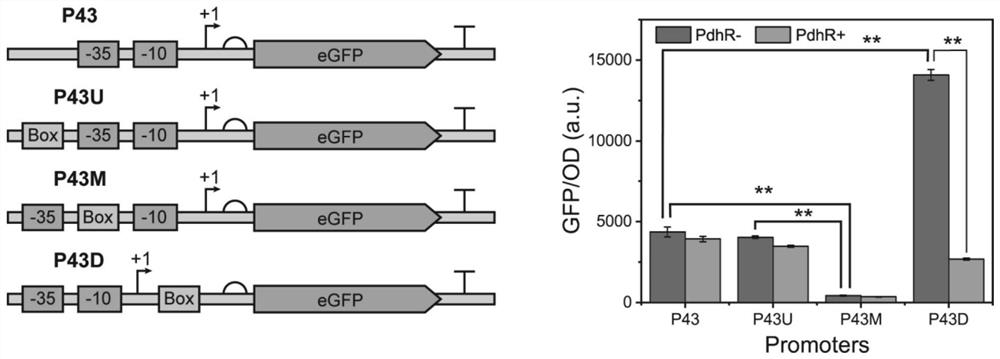
[0046] Using the plasmid PHT01-P43-GFP preserved in the laboratory as a template (containing the P43 promoter, and the reporter gene egfp is placed downstream of P43), P43U-F / P43U-R, P43M-F / P43M-R and P43D-F / P43D-R was used as a primer, and the PdhR-binding sequence was inserted into the -35 region, -10 region and +1 site of the P43 promoter in the plasmid using inverse PCR technology to construct a hybrid promoter regulated by PdhR, thereby obtaining a hybrid promoter containing Recombinant plasmids PHT01-P43U-GFP, PHT01-P43M-GFP, and PHT01-P43D-GFP for pyruvate-responsive biosensors. Subsequently, the recombinant plasmids were transformed into Bacillus subtilis 168 and the recombinant strain 168-PR in Example 1 using the electric shock transformation technique, and then the fluorescence intensity of each recombinant plasmid in the corresponding strain was detected according to the detection method 2. The result is as ...
Embodiment 3
[0047] Example 3: Verification of the binding constant between P43D and pdhR in vitro
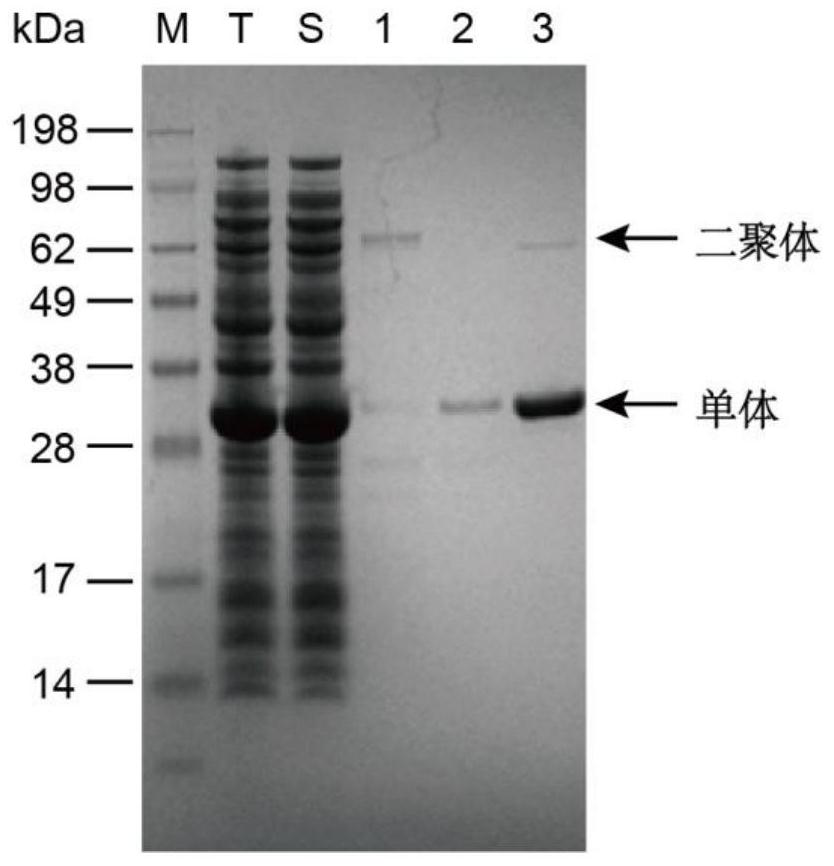
[0048] (1) Use the plasmid pET-28a preserved in the laboratory as a template, Z_PET28F and Z_PET28R as primers, and obtain linearized vector pET-28a by PCR amplification; use the Escherichia coli K12 genome as a template, 28apdhRF and 28apdhRR as primers, and perform PCR amplification A pdhR fragment containing homology arms was obtained. Then use NEB's Gibson Master Mix fused the pdhR fragment with the linearized vector pET-28a (refer to the product manual for specific steps) to obtain the recombinant plasmid pET-28a-pdhR. The recombinant plasmids were sent for sequencing, and the correctly sequenced plasmids were stored at -20°C.
[0049] (2) Transform the recombinant plasmid obtained in step (1) into E.coli BL21 competent (for the operation method, refer to the Basic Molecular Biology Manual), and obtain the recombinant strain E.coli BL21-pdhR containing the plasmid pET-28a-pdhR . T...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com