Cancer precise chemotherapy typing marker screening method, chemotherapy sensitivity molecular typing method and application
A screening method and molecular typing technology, applied in the field of tumor clinical medicine, can solve problems such as unreliable answers, and achieve the effect of long storage time, large quantity, and complete follow-up information
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0080] Example 1 Obtaining protein expression profile data of gastric cancer FFPE samples
[0081] Protein extraction and analysis were performed on 1020 FFPE samples of known gastric cancer cases, and the detailed steps were based on the steps described in the first part of Example 1 in the patent application CN110146636A.
[0082] Perform mass spectrometry detection on the sample, and for detection and analysis, refer to the specific implementation part of the patent application CN108445097A "3. Mass spectrometry detection of gastric cancer protein samples; 4. Mass spectrometry data analysis of gastric cancer protein samples":
[0083] Use the Firmiana computing platform to analyze mass spectrometry data, identify peptides and assemble proteins, and use the iBAQ algorithm for protein quantification to form protein expression profile data, including: protein identifiers, such as Protein GInumber, Accession or Gene Symbol; corresponding protein Quantitative expression values ...
Embodiment 2
[0084] Example 2 Selection of typing markers
[0085] Based on the data in Example 1, the typing markers were selected, and the specific steps were as follows:
[0086] 1) Protein expression profile preprocessing and experimental filtering
[0087] a) High-confidence protein screening: Quantitative proteins are required to contain at least one unique peptide with an ion score greater than or equal to 20, and at least two peptides with an ion score greater than or equal to 20, or three ion scores Peptides with a value greater than or equal to 20.
[0088] b) Eliminate experiments with a protein identification frequency lower than 700 (this step can be adjusted according to the actual number of protein identifications in different cancers and different sample types).
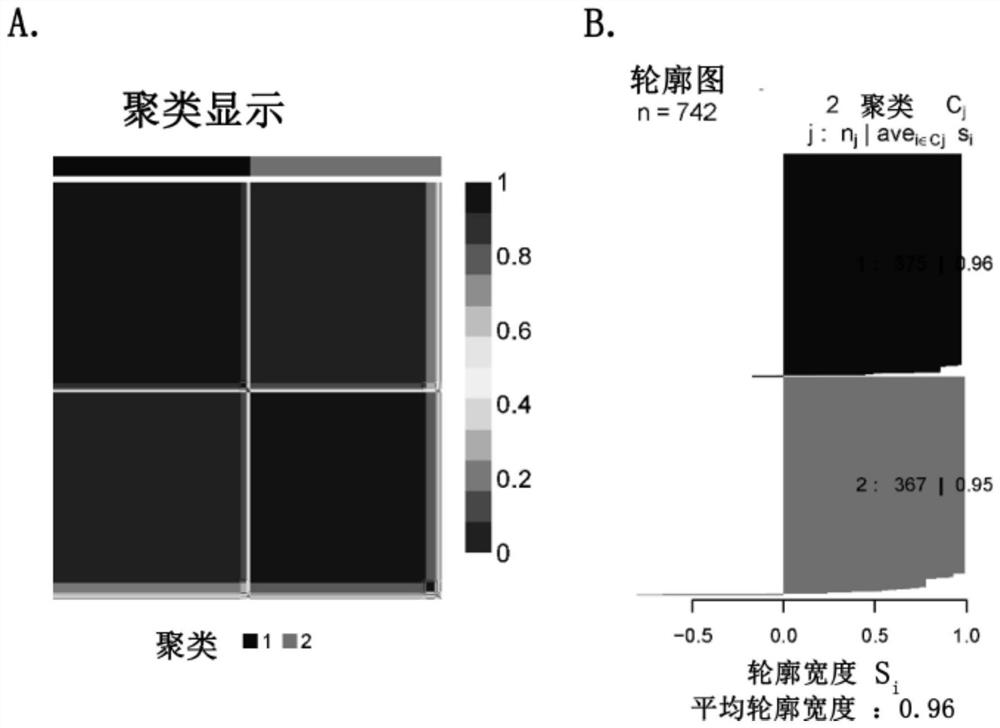
[0089] c) Divide the 1020 samples into a discovery set (742 samples) and a validation set (278 samples).
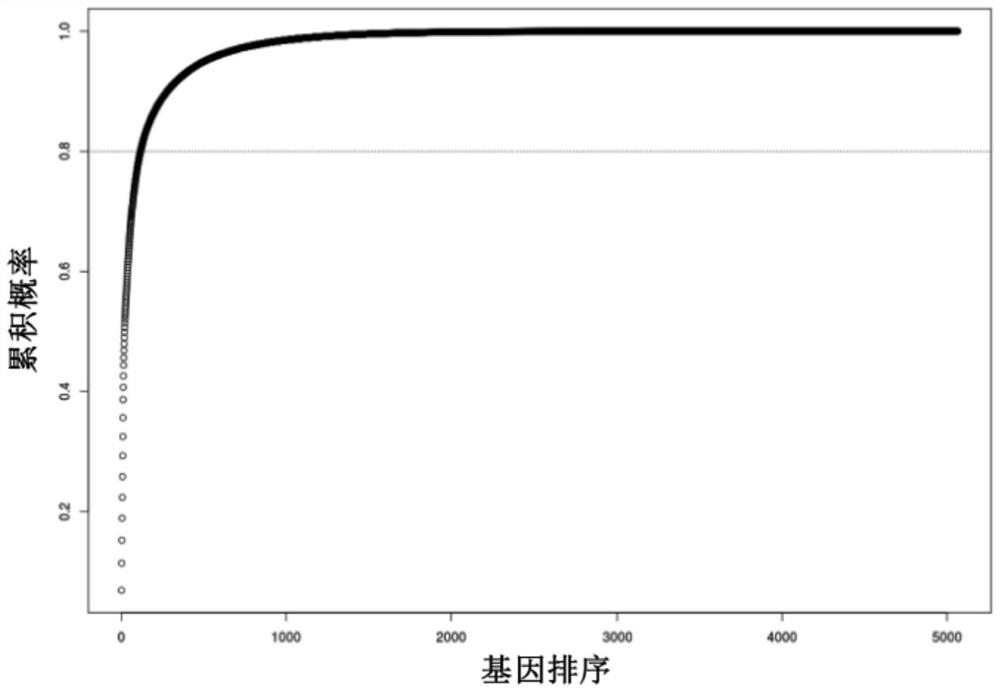
[0090] d) Remove redundant proteins based on the discovery set composed of 742 samples: the sum of 5% p...
Embodiment 3
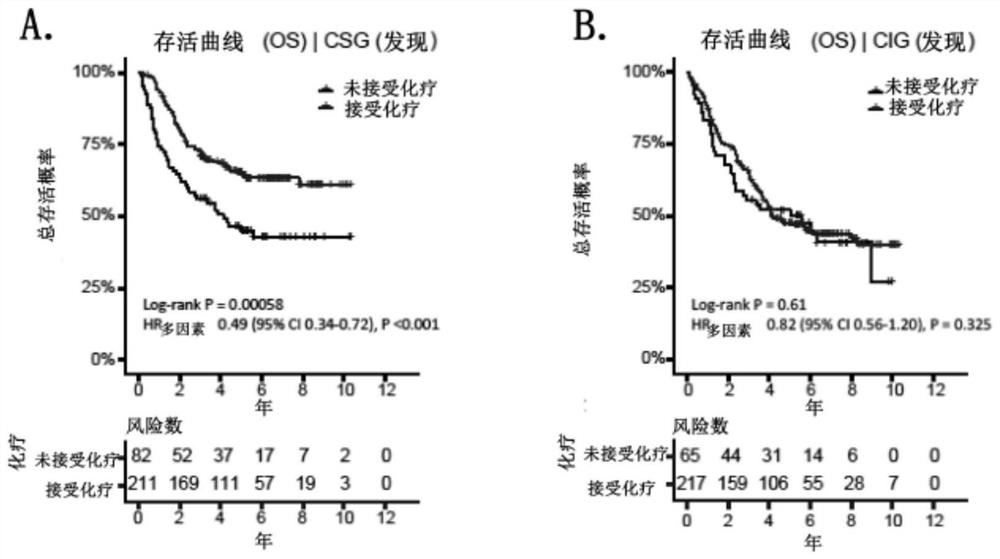
[0101] Example 3 Molecular classification of precision chemotherapy for gastric cancer
[0102] Part I: Molecular classification of precision chemotherapy
[0103] (1) Perform the following typing steps on the typing marker set obtained in Example 2:
[0104] a. Use log(1+x) to convert the FOT value of each protein in the typing marker set, where x is the FOT value of the protein; the converted data are shown in Table 2 below (in the table, the row is typing markers, columns are experiments):
[0105] Table 2
[0106]
[0107] b. Use the non-negative matrix factorization (NMF) consensus clustering method to classify the typing marker set, and assign an NMF typing label to each typing, as follows:
[0108] Load the R language package CancerSubtypes, use the ExecuteCNMF function to analyze the expression profile of the typing markers, set the parameter clusterNum to 2, and set nrun to 50.
[0109] The NMF typing results are shown in Table 3:
[0110] table 3
[0111] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com