Metagenomics-based pathogen analysis method, analysis device, equipment and storage medium
A technology of metagenomics and analysis methods, applied in sequence analysis, informatics, biostatistics, etc., can solve the problems of mNGS standardization obstacles, mNGS experiment repeatability, result reliability and interpretation accuracy, and achieve faster data The effect of analyzing time, improving reference meaning and interpretability, and preventing missed detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
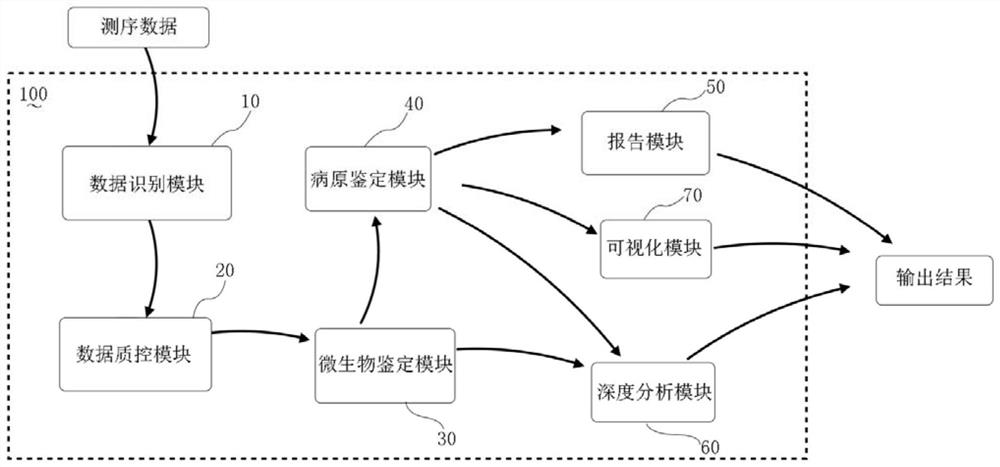
Image
Examples
Embodiment 1
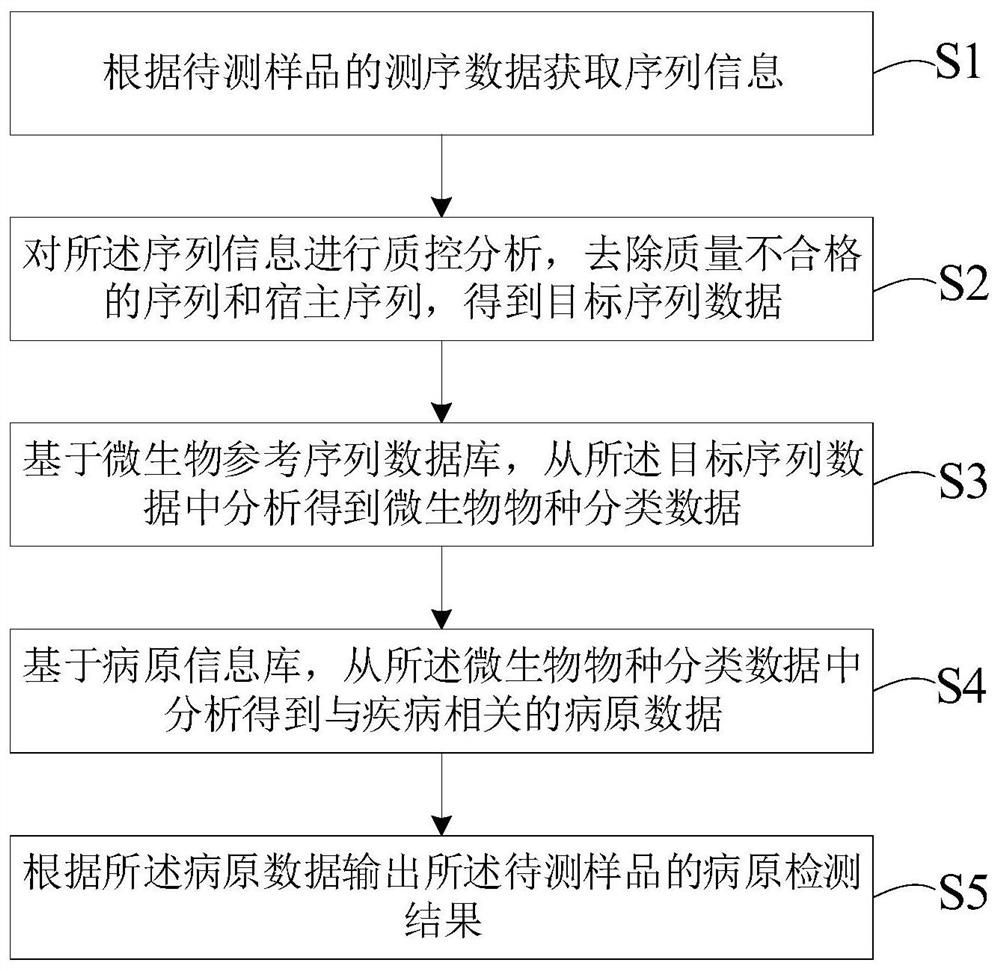
[0126] 1. Metagenome sequencing
[0127] On August 9, 2019, the bronchoalveolar lavage fluid of 5 patients with chronic bronchitis was collected. After microbial nucleic acid extraction, metagenomic sequencing was performed using illumina HiSeq X to obtain off-machine sequencing data.
[0128] First, the update module will update the required database resources, organize and write them into the microbial reference sequence database and the pathogen information database, and record the update information in the local log file. Then, analyze according to the following steps.
[0129] 2. Automatic identification of sequencing data
[0130] The sequencing data is in FASTQ format. Each sequence in the FASTQ format file consists of four lines. The first line starts with the @ symbol, which is the sequence header information, the second line is the base sequence, and the fourth line is the quality value corresponding to each base. A typical FASTQ sequence of the data set of this ex...
Embodiment 2
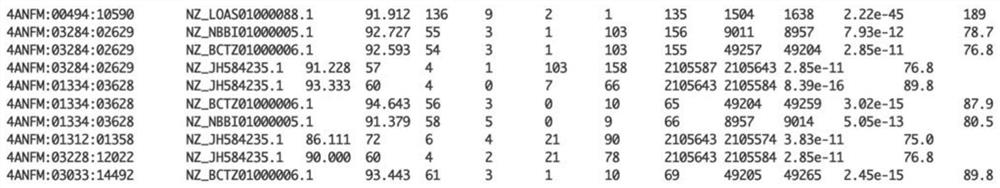
[0159] Such as Figure 17 As shown, using the above-mentioned pathogen analysis method of the present invention and the traditional mNGS method (including the comparison method based on Diamond, the method based on Kraken software and the method based on Kaiju software) to detect a sample of known theoretical microbial composition, the results show that, The analytical method of the present invention is superior in reliability and accuracy of results. In the figure, the horizontal axis represents different detection methods, and the vertical axis represents the abundance of detected species.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com