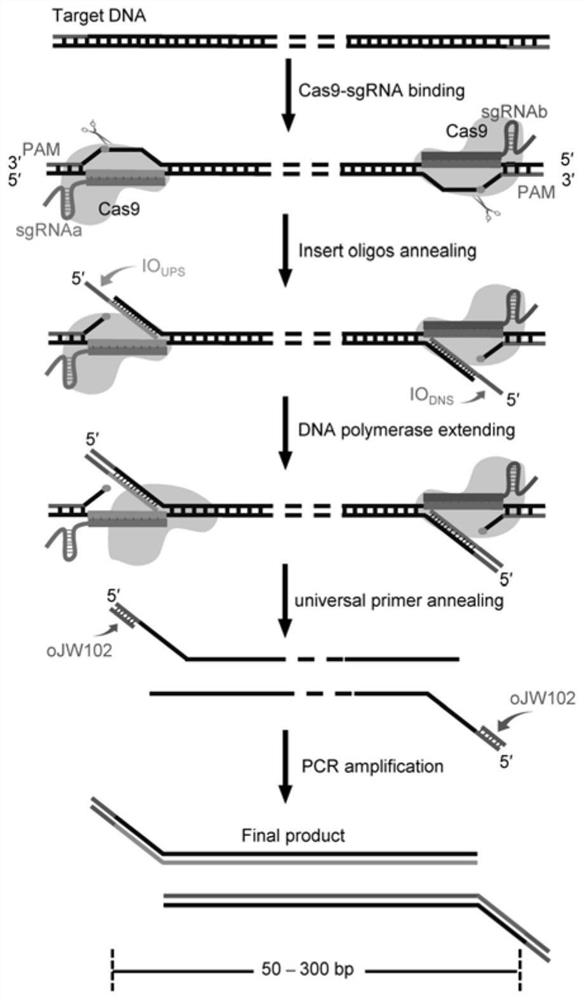
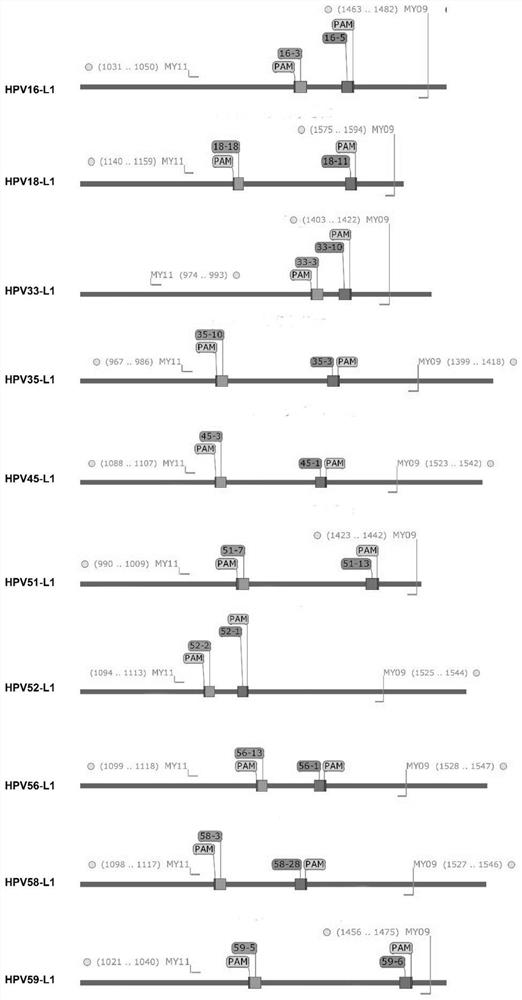
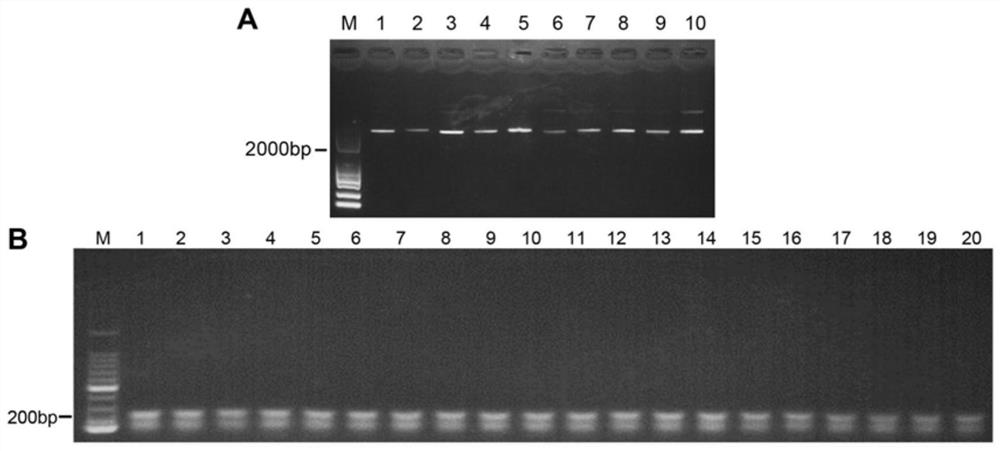
CRISPR/Cas9 typing PCR method for DNA homogeneous detection and application of CRISPR/Cas9 typing PCR method
A detection reagent, a pair of technology, applied in the field of CRISPR/Cas9 typing PCR, can solve the problem of not realizing single-tube homogeneous detection, and achieve the effect of high specificity and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] ctPCR4.0 detection of high-risk HPV DNA in different samples
[0054] 1. Experimental materials and methods
[0055] 1.1. Design of sgRNA
[0056] The sgRNA was designed using the online sgRNA design software Chop-Chop (http: / / chopchop.cbu.uib.no / ), and hg19 was used as the reference genome in the design. Table 1 shows the designed sgRNAs targeting 10 high-risk types of HPV (hrHPV). Two sgRNAs, sgRNAa and sgRNAb, were designed separately for each DNA target. Based on the designed sgRNA, primers for amplifying the sgRNA template were synthesized through a three-round fusion PCR protocol (Table 2). The sgRNA transcription template amplified by PCR has a T7 promoter sequence. The sgRNA templates were then used to prepare sgRNAs by in vitro transcription.
[0057] 1.2. Preparation of sgRNA in vitro transcription template
[0058] PCR1: first design a pair of primers (F1 and R shown in Table 2) according to the backbone part of the sgRNA for fusion PCR amplification. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com