Myasthenia gravis diagnostic reagent kit taking microorganisms as diagnostic markers and application
A myasthenia gravis and microbial technology, applied in the biological field, can solve the problems of treatment delay, easy misdiagnosis as oculomotor nerve paralysis, etc., and achieve the effect of high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
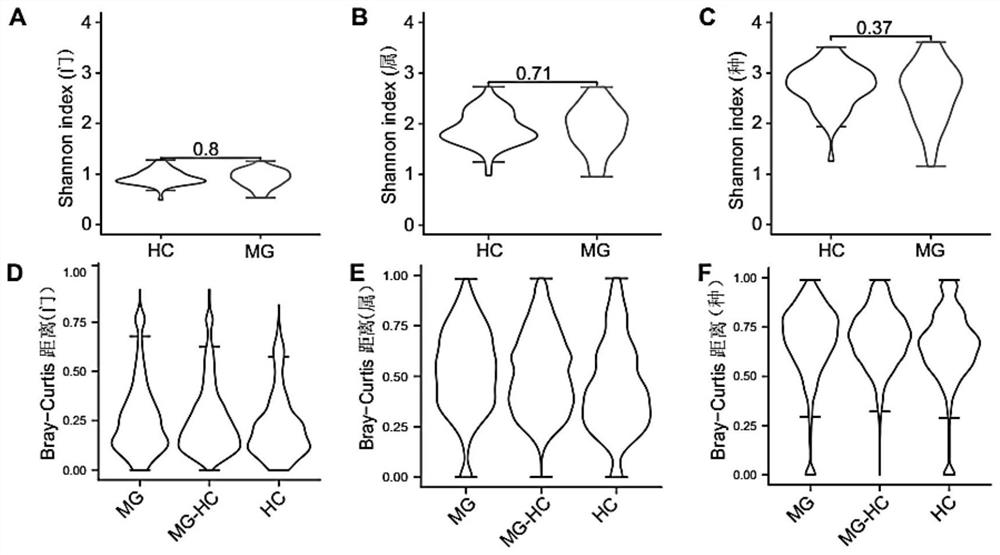
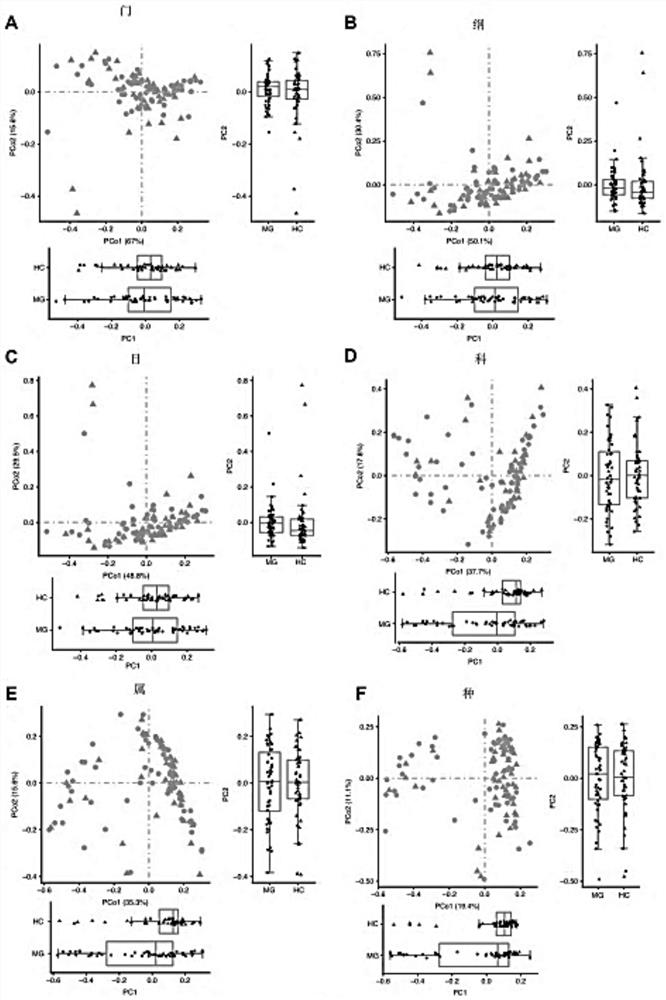
[0053] Example 1 Screening of intestinal flora associated with myasthenia gravis
[0054] 1. Research objects and sample collection
[0055] 55 cases of children with myasthenia gravis and 36 healthy controls (HC) of corresponding age and sex were collected in the Myasthenia Gravis Treatment Center of the First Hospital of Shijiazhuang City, Hebei Province. The sample information is shown in Table 1.
[0056] Diagnostic criteria: (1) clinical manifestations: ptosis, diplopia, strabismus; (2) neostigmine test positive: (3) acetylcholine receptor antibody antibody positive: (4) electromyography: facial nerve attenuation at low frequency, high frequency No increment. Those who meet one of (1)+(2) or (3) or (4) can be diagnosed clearly.
[0057] Type: Refer to the new clinical type and quantitative myasthenia gravis score (QMG) standard type proposed by the Myasthenia Gravis Association of America (MGFA) in 2000.
[0058] Inclusion criteria: The patients were clearly diagnosed...
Embodiment 2
[0085] Example 2 Verifies the accuracy of genome sequencing
[0086] According to the method of Example 1, 19 samples of myasthenia gravis and 13 samples of healthy people were collected, and the patient information is shown in Table 4.
[0087] Table 4 Sample clinical characteristics
[0088]
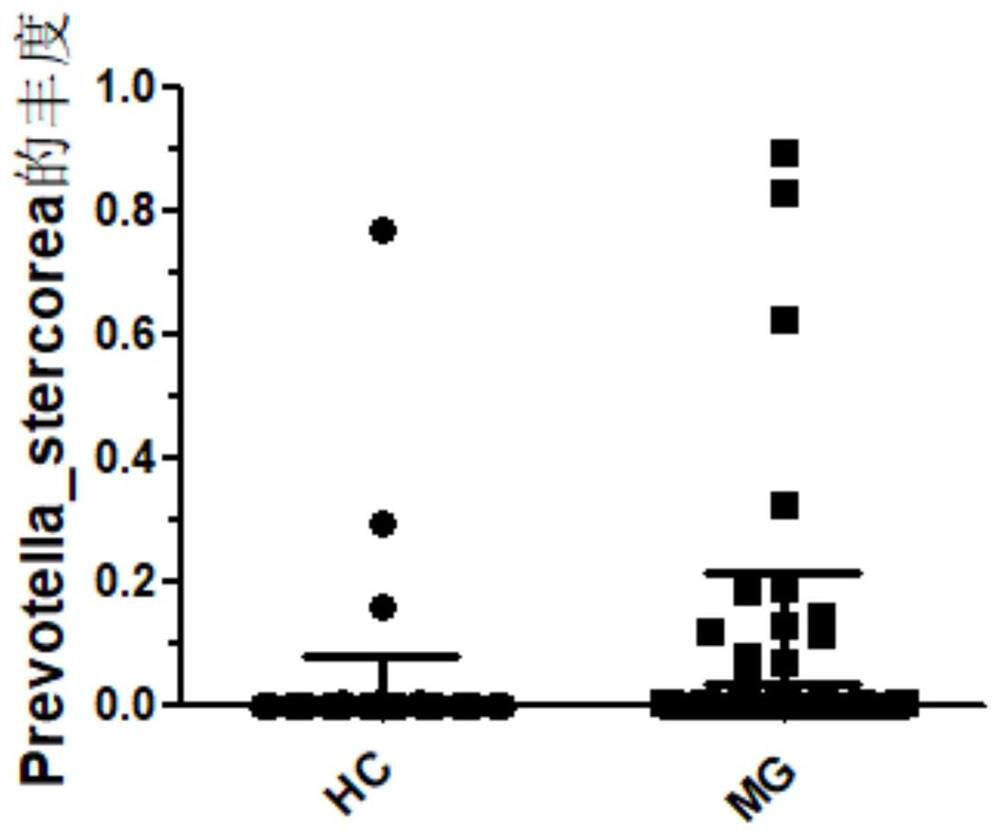
[0089] Differential flora Prevotella_copri, Clostridium_bartlettii, Fusobacterium_mortiferum, and Helicobacter_cinaedi were randomly selected for sequencing verification, and their diagnostic efficacy for myasthenia gravis was calculated.
[0090] The results showed that the AUC values of Prevotella_copri, Clostridium_bartlettii, Fusobacterium_mortiferum, and Helicobacter_cinaedi were 0.736842105, 0.672064777, 0.821862348, and 0.615384615, respectively, which were comparable to the previous detection results, indicating that the sequencing data of the metagenomic group was accurate.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com