A method for analyzing plant endophytic bacterial flora with primers in the v5v6 region
A plant endophytic bacteria, V5-V6 technology, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., can solve problems such as pollution, mitochondrial sequence pollution, interference with flora analysis, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Embodiment 1, analyze the primer design and synthesis of plant endophytic bacterial flora
[0039] In existing studies, 799F is the only primer capable of avoiding plant chloroplast contamination, and the 799F is a single-stranded DNA molecule whose nucleotide sequence is shown in sequence 1 in the sequence listing.
[0040] In order to avoid host plant mitochondrial 18S rDNA in the present invention, primer B is designed according to the V5-V6 region of bacterial 16S rDNA as follows:
[0041] The primer B corresponds to the plant mitochondrial 18S rDNA, and the 3' end of the primer B has 3 consecutive different nucleotides from the plant mitochondrial 18S rDNA, and the full length of the primer B corresponds to the plant mitochondrial 18S rDNA There are 4 or more different nucleotides in the rDNA, except for the different nucleotides in the primer B, other nucleotides are identical or complementary to the corresponding nucleotides of the plant mitochondrial 18S rDNA. ...
Embodiment 2
[0046] Embodiment 2, the establishment of the amplification method of analysis plant endophytic bacterial flora
[0047] Using the total DNA of rice leaves as a template, primers 799F and 1107R (concentration 10 μM) described in Example 1 as primers, Platinum Taq DNA polymerase with hot start activity and no 3'→5' exonuclease correction activity (Invitrogen, USA) is an enzyme, and PCR amplification is performed to obtain a PCR amplification product.
[0048] The reaction system of above-mentioned PCR amplification is as follows:
[0049]
[0050] The flow process of the above-mentioned PCR amplification is as follows:
[0051]
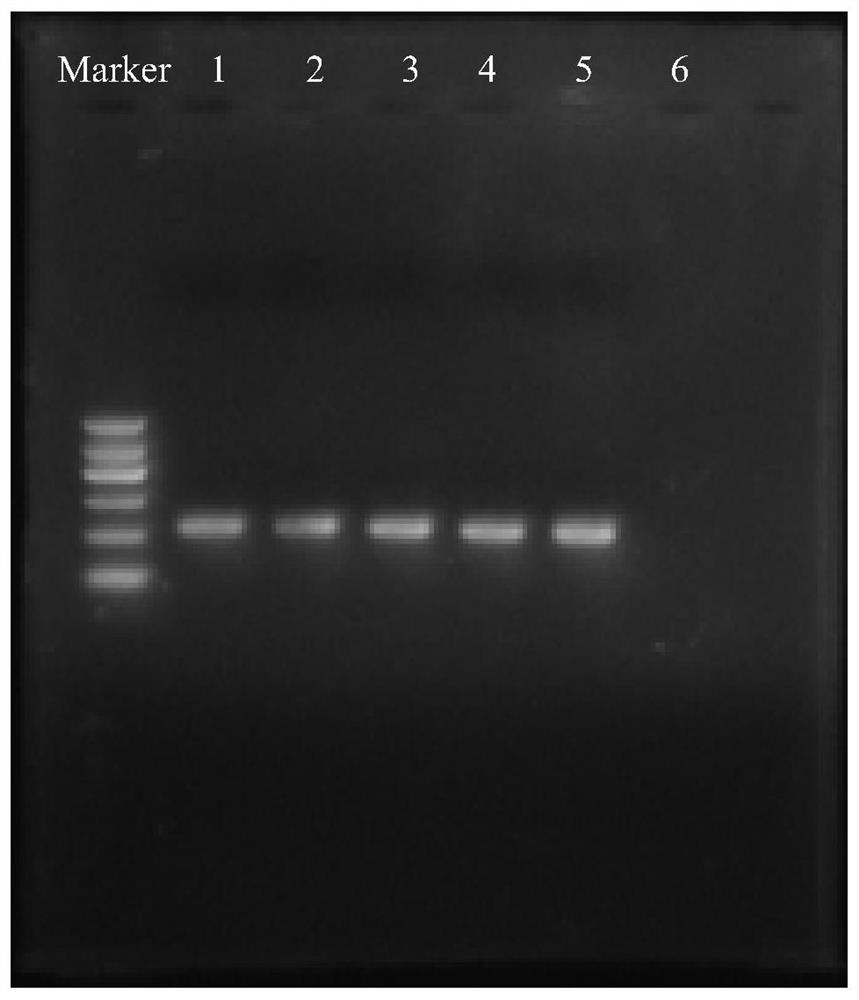
[0052] The PCR amplification products were electrophoresed on 1% agarose gel, and the results were as follows: figure 1 As shown, the amplified product with a length of about 350bp was obtained, which was the V5-V6 region of the 16S rDNA of the bacterial flora.
Embodiment 3
[0053] Example 3, High-throughput sequencing method for analyzing plant endophytic bacterial flora
[0054] 1. PCR amplification:
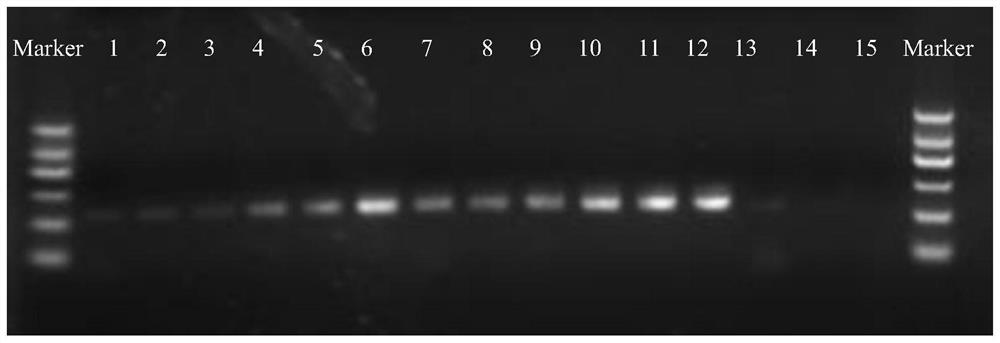
[0055] Using the total DNA from leaves of four plants: citrus (Citrus sinensis, Cr), tomato (Lycopersicon esculentum, Le), Arabidopsis thaliana (At), rice (Oryza sativa, Os) as templates, each plant Set up 3 samples, and add 6 base lengths to the 5' ends of the primers 799F and 1107R synthesized in Example 1 to distinguish the Barcode of different samples. The primer Barcode combination of each sample is different for subsequent high-pass The sample identification of quantity sequencing; Utilize the amplification method of embodiment 2 to carry out PCR amplification, each sample obtains the amplified product of 150 μ l, amplified product is electrophoresed with 1% agarose gel, and the result is as follows figure 2 shown.
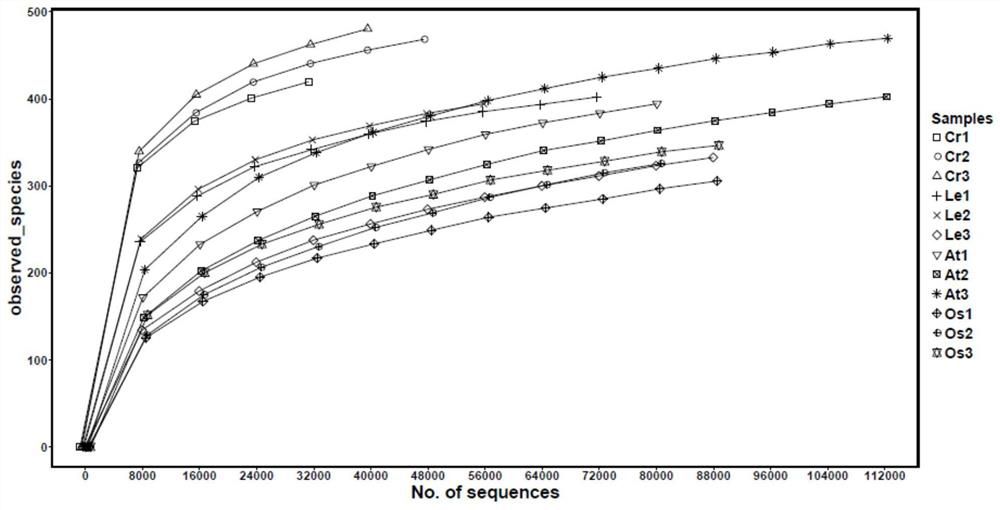
[0056] 2. Illumina sequencing results
[0057] Using the Illumina Hiseq 2500 sequencing platform of Guangdong Meige Gene T...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com