Method for detecting prostate cancer exosome based on Fe3O4@ SiO2@ TiO2 nanoparticle enrichment and PSMA sensor
A nanoparticle, prostate cancer technology, applied in instruments, measuring devices, scientific instruments, etc., can solve problems such as loss of exocytosis, and achieve the effects of strong specificity, simple and rapid separation method, and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Synthetic Fe 3 o 4 @SiO 2 @TiO 2 Nanoparticles
[0048] 1. Add 0.15g Fe 3 o 4 Dissolve in a mixture of ethanol (280mL), deionized water (70mL) and ammonia water (5mL, 28wt%), and sonicate for 15min. Then 4 mL TEOS was added, and the reaction was continuously stirred at room temperature for 10 h to obtain Fe 3 o 4 @SiO 2 . figure 1 a is Fe 3 o 4 TEM image of figure 1 b is Fe 3 o 4 @SiO 2 TEM image. These two figures illustrate that SiO 2 Successfully wrapped in Fe 3 o 4 Outside.
[0049] 2. Add 0.05g Fe 3 o 4 @SiO 2 Dissolve in a mixture of ethanol (100 mL) and ammonia water (0.3 mL, 28 wt%), and after ultrasonication for 15 min, add 0.75 mL of TBOT dissolved in ethanol dropwise with continuous stirring. The reaction was continuously stirred at 45° C. for 24 h, the prepared product was washed three times with ultrapure water and ethanol, and dried at 60° C. overnight. figure 1 c is Fe 3 o 4 @SiO 2 @TiO 2 TEM image of figure 1 d is Fe 3 o 4 ...
Embodiment 2
[0051] Exosome isolation.
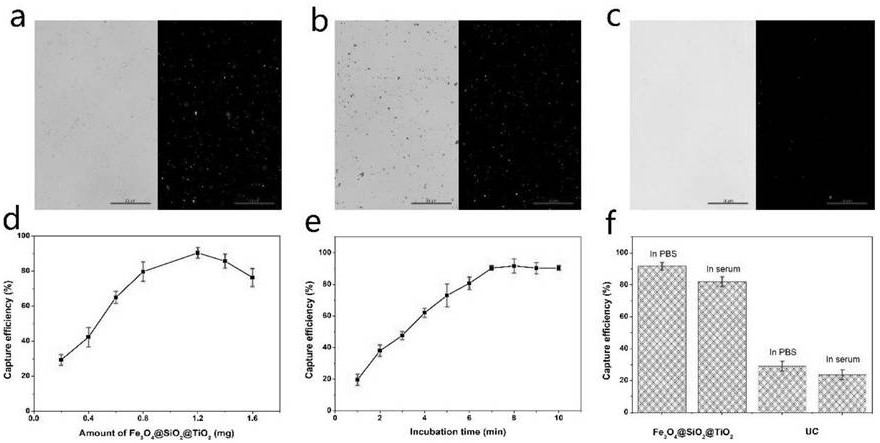
[0052] 1. Prepare two parallel samples: LNCaP exosome solution (10 7 individual / μL). Exosomes from one sample were stained with PKH26 dye. Exosomes from another sample were stained with 1.2 mg Fe 3 o 4 @SiO 2 @TiO 2 After incubating at room temperature for 8 minutes, use a magnetic stand for rapid magnetic separation, and wash with PBS repeatedly for 3 times. figure 2 a is the bright-field and fluorescence images after exosome staining, figure 2 b is Fe 3 o 4 @SiO 2 @TiO 2 / bright-field and fluorescence maps of exosomes, figure 2 c is the bright-field and fluorescence images of the washings after repeated washing. These three figures illustrate that Fe 3 o 4 @SiO 2 @TiO 2 Exosomes can be successfully isolated and the number of enriched exosomes is large.
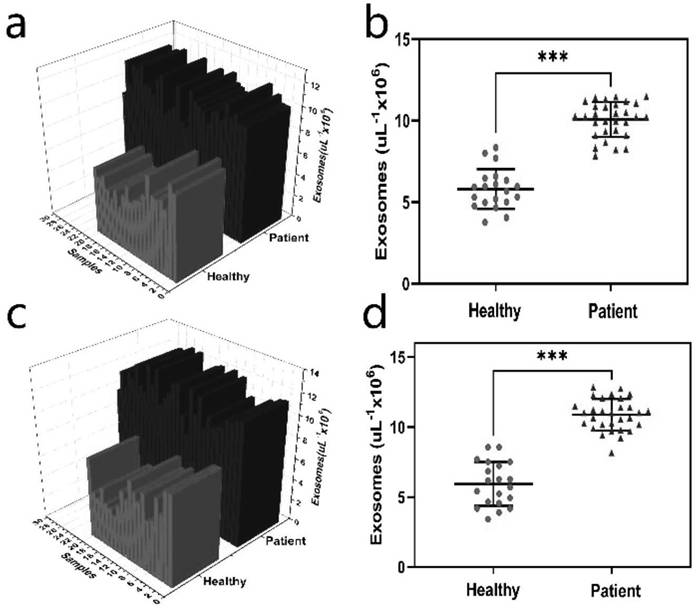
[0053] 2. Prepare exosomes at a concentration of 10 7 cells / μL, stained with PKH26, and the fluorescence intensity was recorded as F 0 . Prepare the same concentration of ex...
Embodiment 3
[0056] Example 3 Exploration of the best conditions for exosome detection, including the following steps:
[0057] 1. Optimization of PSMA sensor. First design and build three PSMA sensors, the PSMA1 sequence is shown in the table below:
[0058]
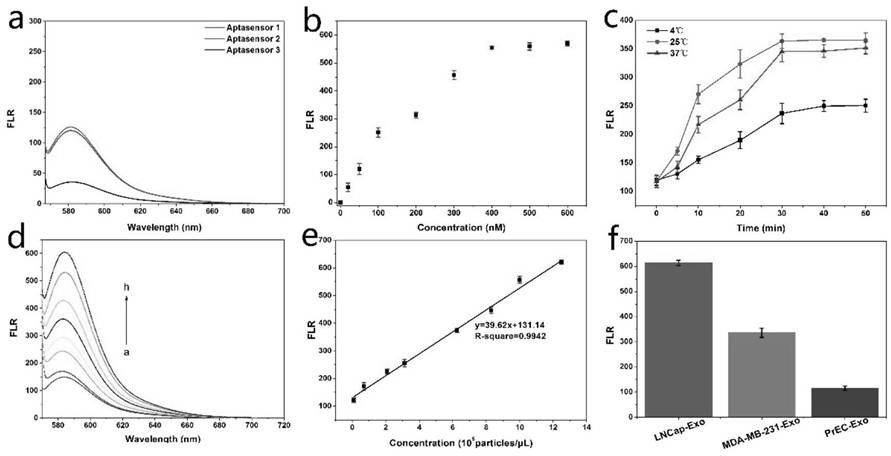
[0059] After synthesis, the chains were untied at a high temperature of 100 degrees, then annealed, and then the three sensors were diluted to a concentration of 200nM and their fluorescence intensity was measured (excitation: 557nm, emission 580nm). image 3 a is the fluorescence intensity graph of three sensors with the same concentration, among which sensor 3 has the smallest fluorescence intensity and the smallest background, so sensor 3 was selected for exosome detection in the experiment.
[0060] 2. Prepare exosomes with a concentration of 106 / μL, incubate with sensor 3 at different concentrations (0, 20, 50, 100, 200, 300, 400, 500, 600nM) at room temperature for 30 minutes, and measure the fluorescence intensity (excit...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com