Molecular markers, detection primers and detection methods for identifying Lactobacillus plantarum and Lactobacillus pentosus
A technology of Lactobacillus pentosus and Lactobacillus plantarum, applied in biochemical equipment and methods, measurement/testing of microorganisms, DNA/RNA fragments, etc., to achieve the effects of reduced detection cost, high specificity, and short detection cycle
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
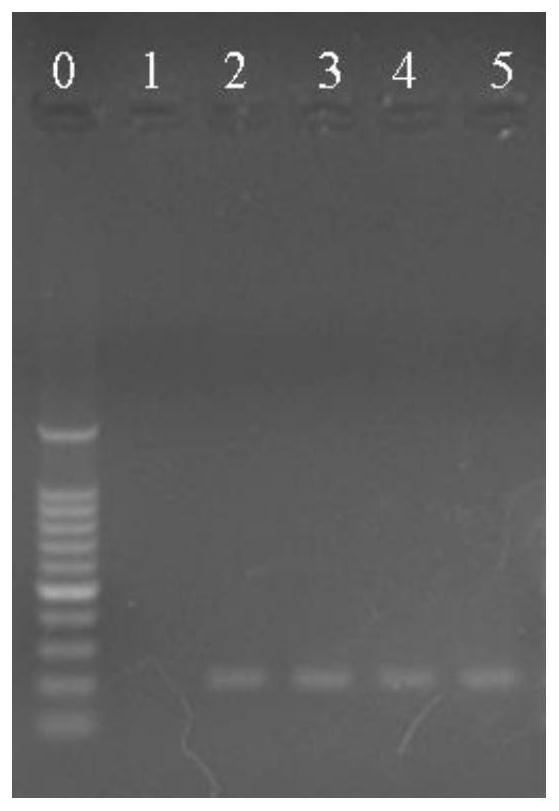
[0059] Embodiment 1: Identification of Lactobacillus plantarum
[0060] 1. Bacterial DNA extraction
[0061] Take 1 mL of the bacterial culture and centrifuge at 12000r / min for 5min, discard the supernatant, collect the bacteria, add 50μL of the lysate of the bacterial DNA extraction kit, suspend and mix well, place in a water bath at 100°C for 15min, ice bath for 3min, and centrifuge at 12000r / min After 10 minutes, the supernatant was taken for later use, and the supernatant contained bacterial DNA as template DNA.
[0062] 2. PCR amplification
[0063] PCR uses a 25 μL reaction system, in which 1.0 μL of the upstream and downstream specific primers of the target DNA at a concentration of 10 μmol / L, 0.5 μL of 1.5U Taq DNA polymerase, and 25 mmol / L MgCl 2 2 μL, 2 μL of 10 mmol / L dNTP, 2.5 μL of 10×PCR buffer, 2.0 μL of DNA template, and 25 μL of sterile deionized water. PCR amplification conditions were: 95°C pre-denaturation for 5 min; 95°C denaturation for 1 min, 58°C an...
Embodiment 2
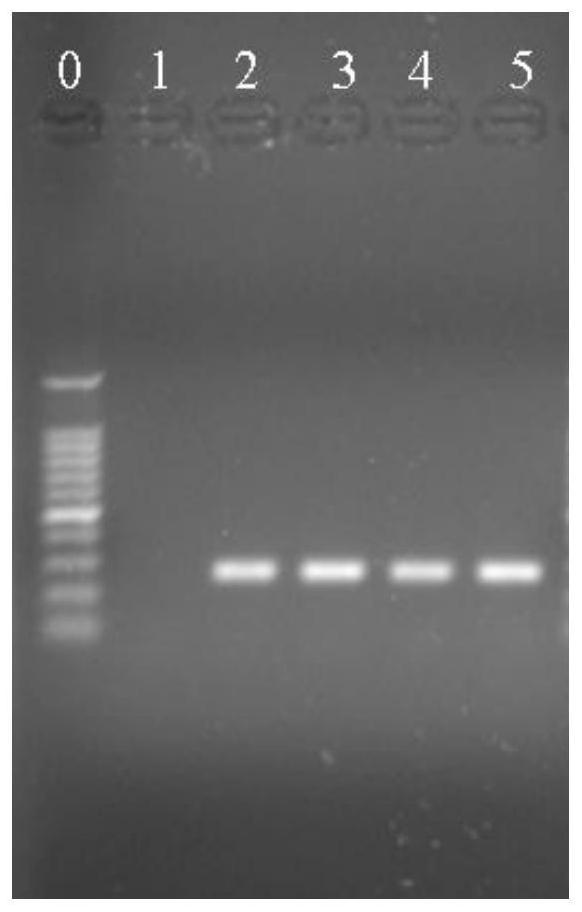
[0066] Example 2: Identification of Pentobacter
[0067] 1. Bacterial DNA extraction
[0068] Take 1 mL of the bacterial culture and centrifuge at 12000r / min for 5min, discard the supernatant, collect the bacteria, add 50μL of the lysate of the bacterial DNA extraction kit, suspend and mix well, place in a water bath at 100°C for 15min, ice bath for 3min, and centrifuge at 12000r / min After 10 minutes, the supernatant was taken for later use, and the supernatant contained bacterial DNA as template DNA.
[0069] 2. PCR amplification
[0070] PCR uses a 25 μL reaction system, in which 1.0 μL of the upstream and downstream specific primers of the target DNA at a concentration of 10 μmol / L, 0.5 μL of 1.5U Taq DNA polymerase, and 25 mmol / L MgCl 2 2 μL, 2 μL of 10 mmol / L dNTP, 2.5 μL of 10×PCR buffer, 2.0 μL of DNA template, and 25 μL of sterile deionized water. PCR amplification conditions were: 95°C pre-denaturation for 5 min; 95°C denaturation for 1 min, 58°C annealing for 45 ...
Embodiment 3
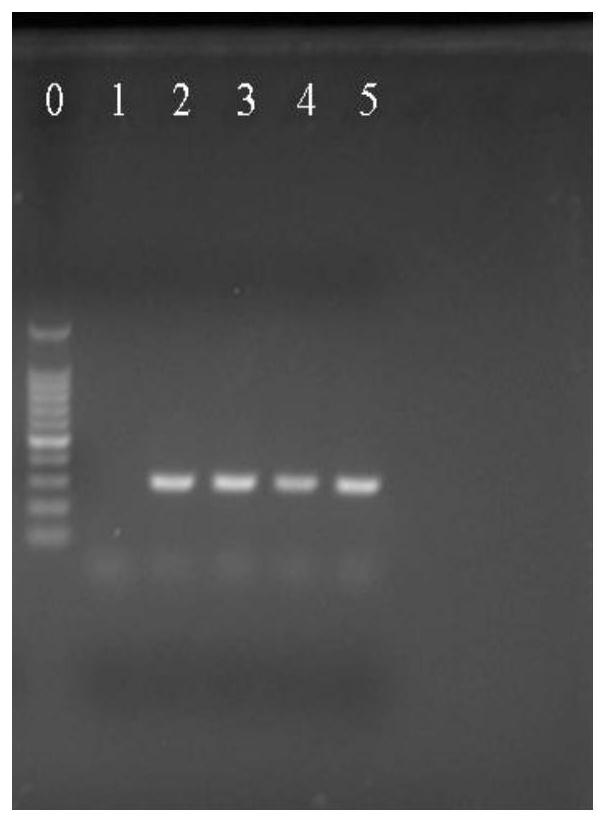
[0073] Example 3: Identification of Lactobacillus plantarum and Lactobacillus pentosus in actual samples
[0074] 1. Sample processing
[0075] For cheese and yogurt samples sold in the market, take 1-2g and add them to 10mL MRS broth, incubate at 37°C for 48h, take 1mL of bacterial liquid, collect the bacterial cells by centrifugation, add 50μL of DNA lysate, suspend and mix well, and place in a water bath at 100°C for 15min , ice-bathed for 3 minutes, centrifuged at 12,000 r / min for 10 minutes, and the supernatant was taken as template DNA. In addition, an inoculation loop was used to streak and inoculate the MRS plate, cultured at 37°C for 48 hours, and a single colony was taken for DNA extraction, PCR identification and mass spectrometry identification.
[0076] 2. PCR amplification
[0077] PCR uses a 25 μL reaction system, in which 1.0 μL of the upstream and downstream specific primers of the target DNA at a concentration of 10 μmol / L, 0.5 μL of 1.5U Taq DNA polymerase, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com