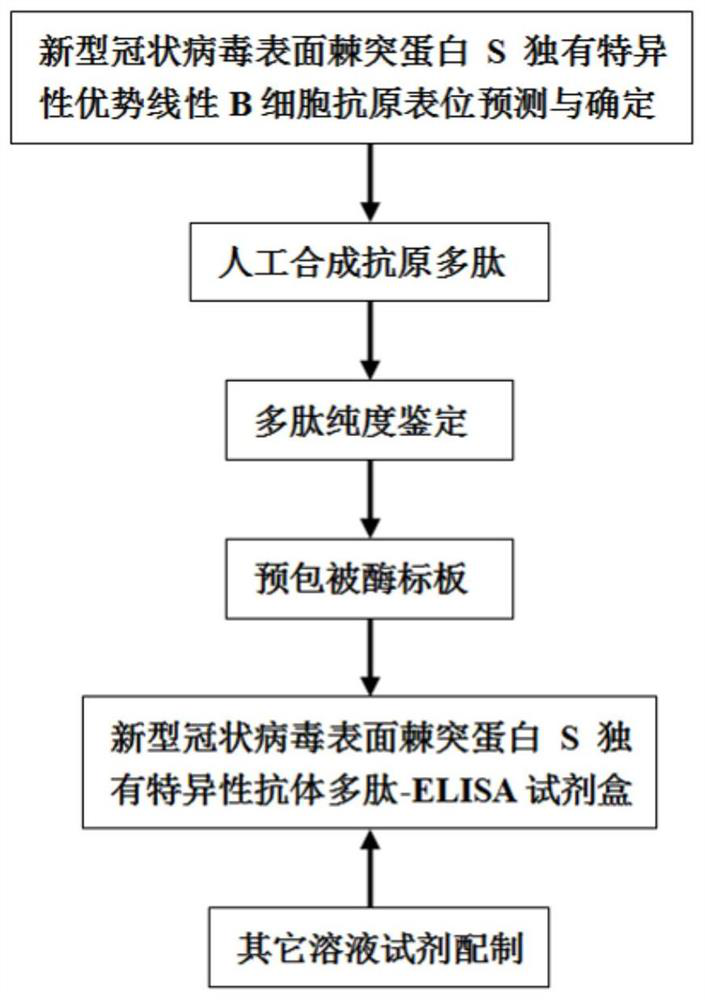
Polypeptide-ELISA kit for detecting novel coronavirus S protein unique antibody
A coronavirus and kit technology, applied in the field of peptide-enzyme-linked immunosorbent assay kits, can solve problems such as affecting specificity, and achieve the effects of simple operation, strong specificity, and improved detection accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
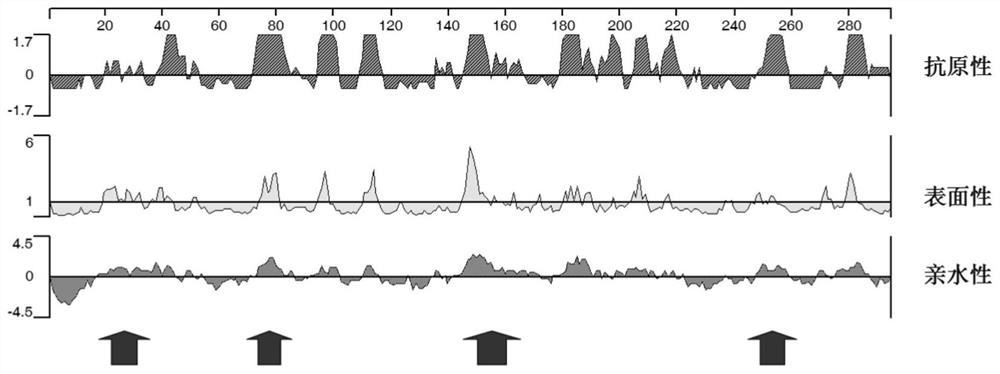
[0032] Example 1: Prediction and determination of the unique dominant linear B cell epitope in the spike protein S on the surface of the novel coronavirus
[0033] According to the characteristics of strong antigenicity, good surface and strong hydrophilicity of the linear B cell epitope, the amino acid sequence of the spike protein S on the surface of the new coronavirus was analyzed using bioinformatics software, and the dominant linear B cell antigen was predicted and determined. Determinant site ( figure 2 ). Use bioinformatics software to compare the amino acid sequences of the surface spike protein S of the new coronavirus SARS-CoV2 with 6 other coronaviruses known to infect humans and cause related diseases, especially the SARS-CoV with the closest homology , to clarify the unique specificity of the above dominant linear B cell epitopes. The peptides were artificially synthesized, and then diluted to 10 micrograms / ml, and coated in a 96-well microtiter plate with an ...
Embodiment 2
[0034] Example 2: Preparation of unique advantage linear B-cell antigen polypeptide on the surface of novel coronavirus spike protein S
[0035] Artificially synthesized four-segment polypeptides (SEQ ID NO.1~SEQ ID NO.4) containing unique dominant linear B cell epitope sites in the surface spike protein S of the novel coronavirus: SQCVNLTTRTQLPPAYTNSFT, HVSGTNGTKRFD, LGVYYHKNNKSWMESEFRVYSS, ALHRSYLTPGDSSSGWTAG, after purification The purity is above 95%.
Embodiment 3
[0036] Example 3: Using the kit to detect the specific antibody of the novel coronavirus surface spike protein S in the sample
[0037] Sample incubation: Take out the microtiter plate in the kit that has been coated with the linear B cell antigen polypeptide with unique specificity and advantages of SARS-CoV2 surface spike protein S. Dilute each sample 100 times with the sample diluent, add to the pre-prepared microplate wells, 100 microliters per well, set up negative control wells, positive control wells and blank wells at the same time, incubate at 37°C for 1 hour, wash with washing solution Plate 3 times.
[0038] Secondary antibody incubation: Dilute the enzyme-labeled antibody 5000 times with sample diluent, add to the wells, 100 microliters per well, incubate at 37°C for 30 minutes and wash the plate 3 times.
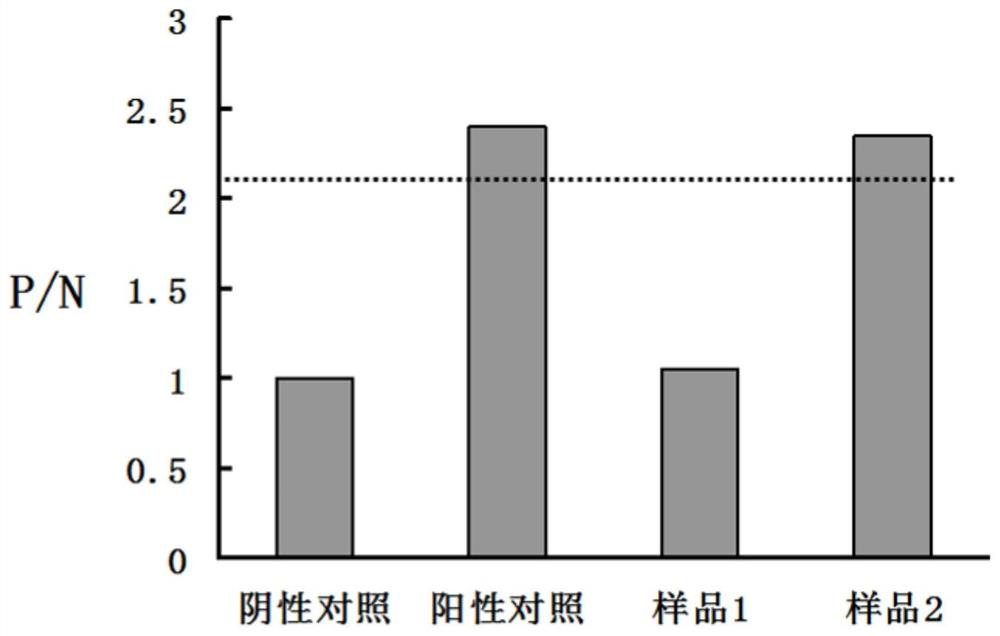
[0039] Color development reading: configure OPD color development solution, weigh 20 mg of enzyme substrate C, dissolve in 50 ml of enzyme substrate A, add 20 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com