Biotin labeled primer extension method and application thereof
A technology of biotin labeling and markers, applied in the field of biochemistry, can solve the problems of low sensitivity of isotope labeling, achieve the effects of increasing storage difficulty, solving safety problems, and optimizing experimental conditions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
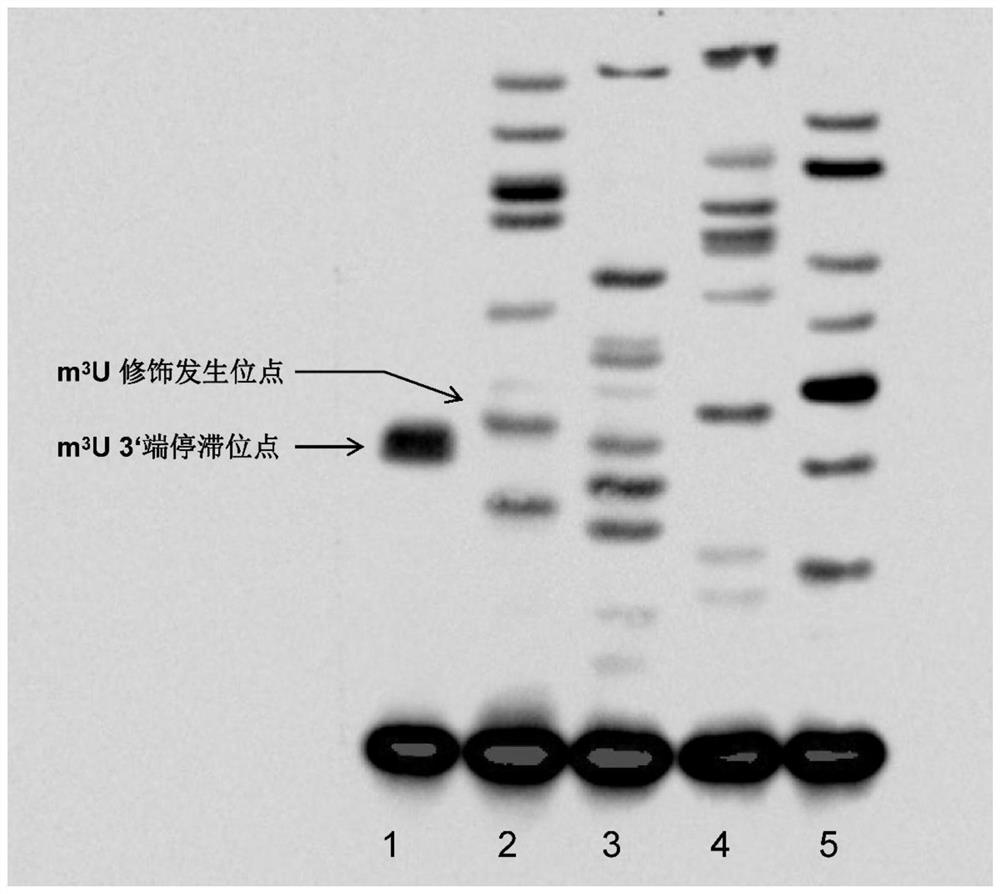
[0086] Example 1 (Saccharomyces cerevisiae RNA m 3 U modification detection)
[0087] Implementation principle:
[0088] The primer extension method utilizes m 3 m 3 U modification for localization detection. m 3 The U modification occurs on the No. 3 N atom of the ribonucleotide guanosine base. This modification replaces the key H atom connected to the N atom for base pairing to form a hydrogen bond with a methyl group (-CH 3 ), hindering the base-pairing process of the guanosine. Taking advantage of this feature, the target RNA sequence is reverse-transcribed using biotin-labeled primers, m3U modification affects the base pairing of its own sites, and part of the cDNA synthesis will be stagnant at m3U 3 The first nucleotide site at the U 3' end forms a stalled cDNA product. Finally, the stalled cDNA product is displayed by gel electrophoresis, membrane transfer, and development. The single-stranded cDNA product prepared by the sanger sequencing method is used as a mark...
Embodiment 2
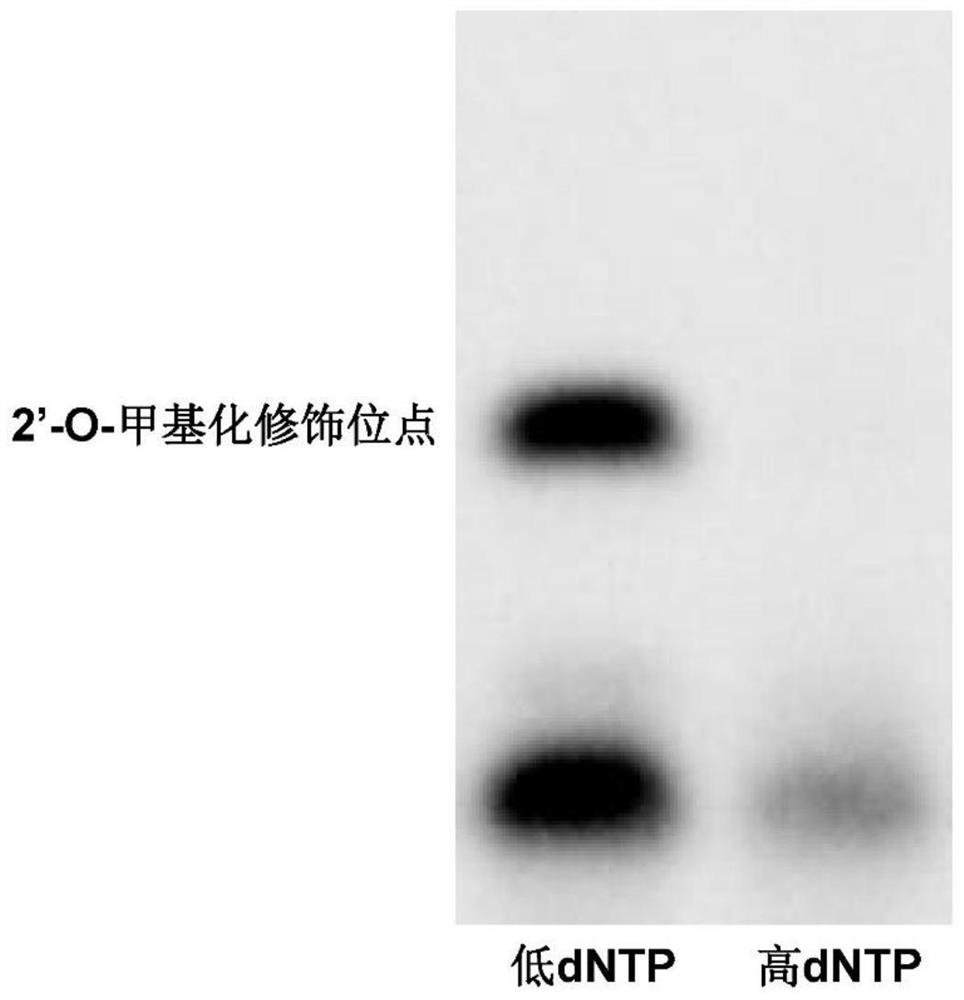
[0105] Example 2 (Drosophila RNA 2'-O methylation modification detection)
[0106] Implementation principle:
[0107] During primer extension, the 2'-O-methylation modification on RNA has a blocking effect on reverse transcriptase, which is mainly affected by the concentration of dNTP. Under the condition of high dNTP concentration, 2'-O-methylation has a weak effect on reverse transcriptase inhibition, while under the condition of low dNTP concentration, 2'-O-methylation has a weak effect on reverse transcriptase inhibition. Large, partially stalled cDNA products are formed during reverse transcription, and these stalled products can be visualized by biotin labeling and gel electrophoresis to determine the site of 2'-O-methylation modification.
[0108] Implementation steps:
[0109]1. Select 20 adult fruit flies (Drosophila melanogaster), freeze them in liquid nitrogen, grind them into pieces, and use the TRIZOL method to extract the total RNA of the flies.
[0110] 2. Sy...
Embodiment 3
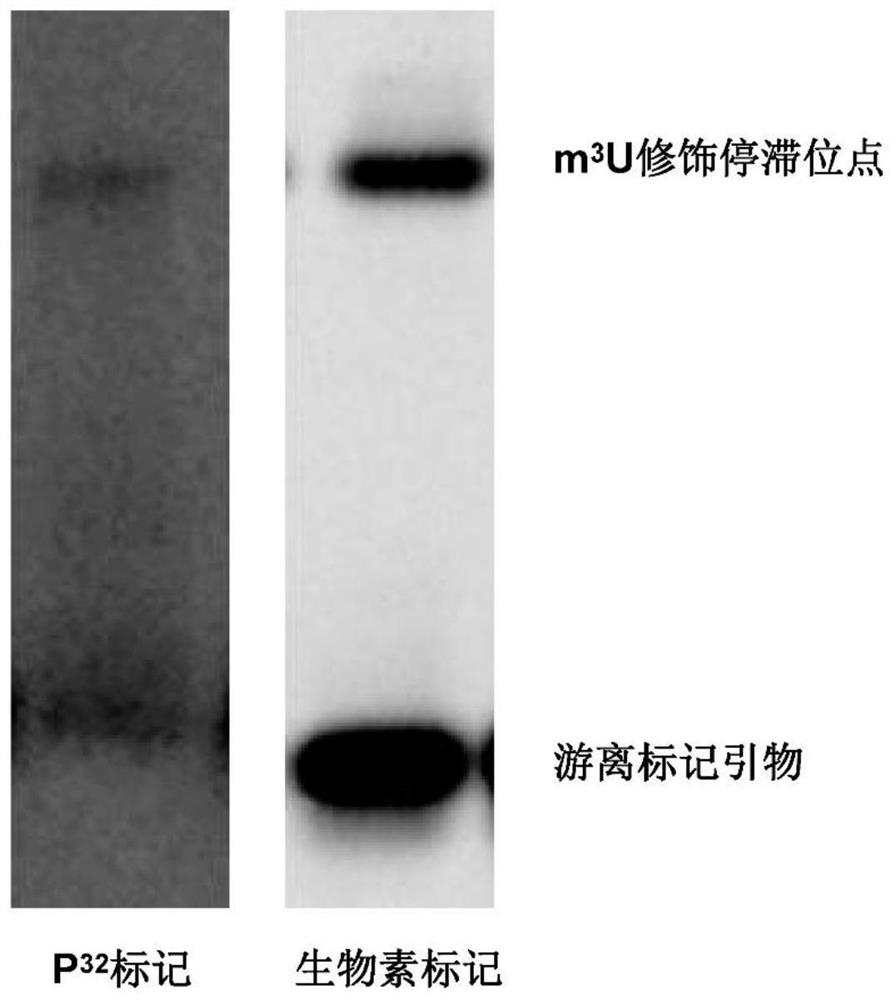
[0119] Example 3: Comparison of development accuracy between biotin primer extension method and traditional isotope method
[0120] One, according to the method of embodiment 1, use biotin-labeled primer (5'-GGTAAAACTAACCTGTCTCA-3') to carry out 28S RNA on the Drosophila total RNA sample 3 U modification site detection, the result is as follows image 3 shown.
[0121] 2. Use P32 isotope-labeled primer (5'-GGTAAAACTAACCTGTCTCA-3') to perform 28S RNA on Drosophila total RNA samples 3 U modification site detection. Specific steps are as follows:
[0122] (1) Reagent formula related to isotope-labeled primer extension method (25 μL system)
[0123] A. Reaction solution A
[0124] Primer (1pmol / μL) 2μL
[0125] Total RNA (2000ng / μL) 4μL
[0126] DEPC treated water 4μL
[0127] B. Reaction solution B (using Promega AMV reverse transcriptase, catalog number: M5101)
[0128]
[0129] (2) Implementation steps of the isotope-labeled primer extension method (all operations a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com