Construction method of prognosis evaluation model of intrahepatic cholangiocarcinoma patient
A technology for prognostic assessment and method construction, applied in the field of bioinformatics, can solve problems that have not yet been
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] Example 1: Data Collection
specific Embodiment approach
[0061] This embodiment mainly introduces the specific steps of collecting DNA methylation data of ICC samples. The specific implementation is as follows:
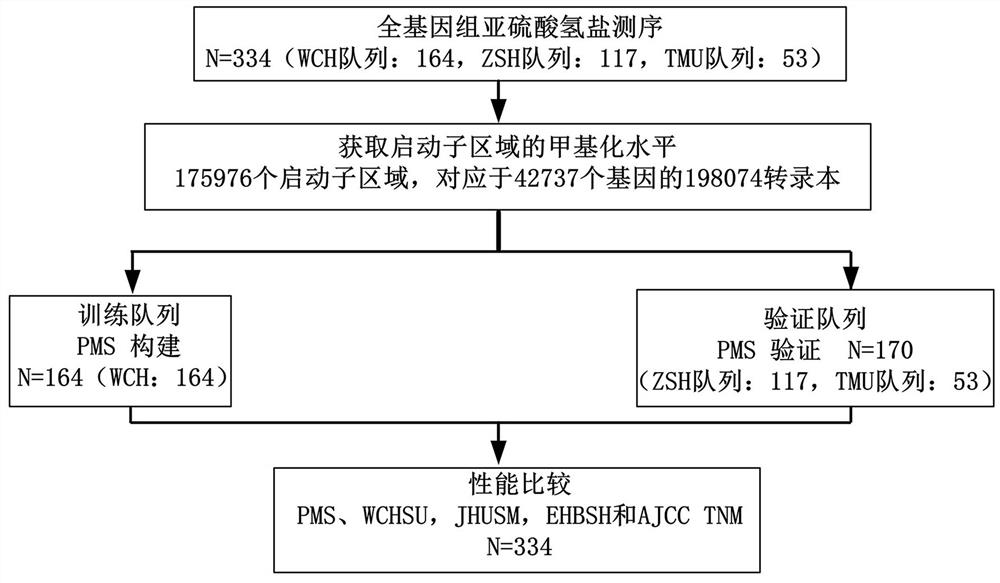
[0062] First, samples of patients with intrahepatic cholangiocarcinoma (ICC) were obtained from the biobanks of West China Hospital of Sichuan University, Zhongshan Hospital of Fudan University, and Tianjin Medical University Cancer Hospital. Immediately after surgery, the samples were placed in liquid nitrogen. Freeze and store at -80 °C or in liquid nitrogen. Then, the samples undergo the process of genomic DNA isolation, WGBS database generation, sequencing, quality control and WGBS data processing to obtain the DNA methylation data of ICC samples. The DNA methylation data include the average methylation value of the 4,000 bps promoter region.
[0063] It should be noted that, as shown in Table 1, 164 and 170 patients participated in the training cohort and the verification cohort respectively in the embodiment of the ...
Embodiment 2
[0070] Example 2: Data Analysis
[0071] Example 2 is mainly to analyze the DNA methylation data in Example 1, and screen out several ICC-related promoter regions. The specific implementation is as follows:
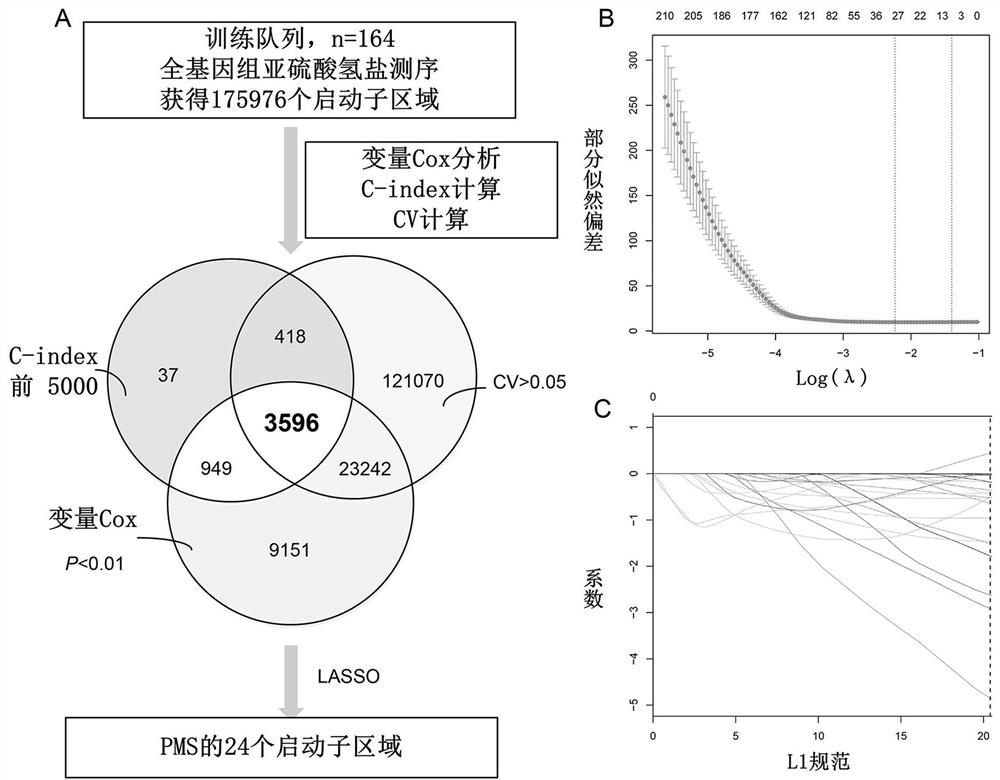
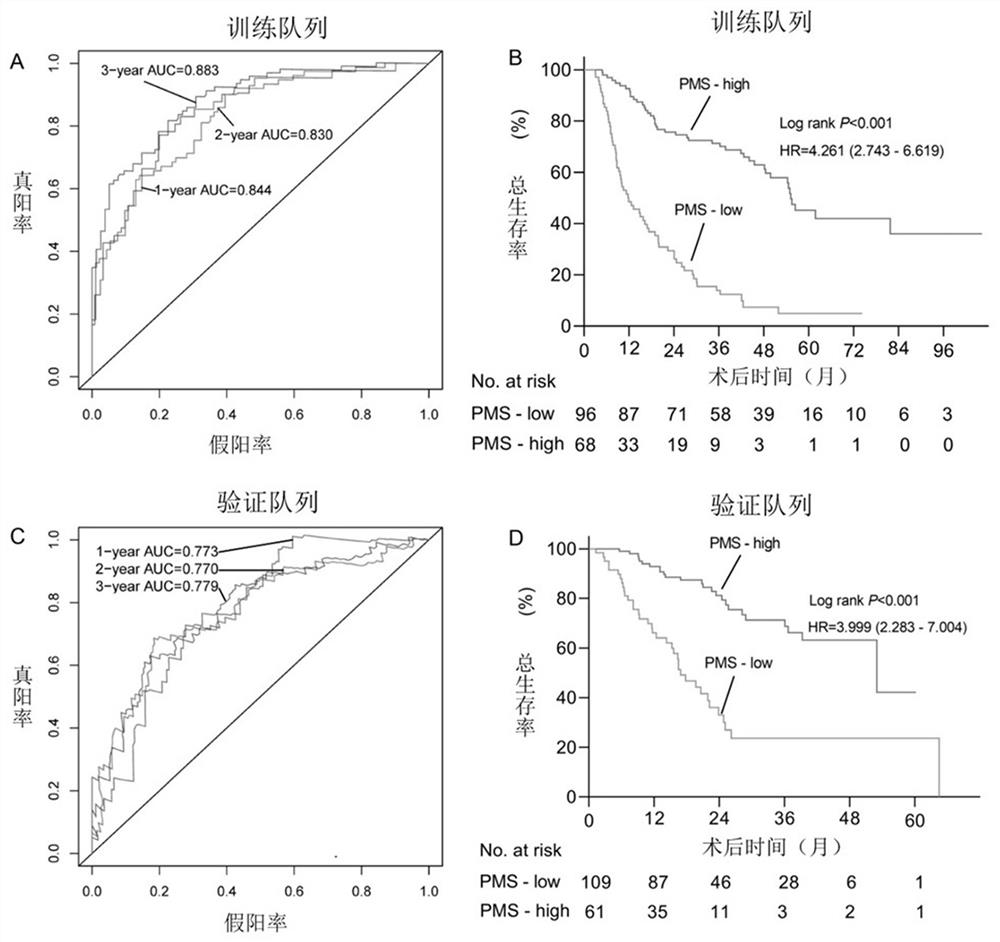
[0072] In total, this example identified 175,976 promoter regions obtained by WGBS in the training cohort, corresponding to 198,074 transcripts for 42,737 genes (coding and non-coding genes). The methylation level of each promoter region derived from the WGBS data was continuous and quantitative, and the methylation level of the promoter region was determined to be significantly associated with overall survival using a univariate Cox method (P < 0.01). Subsequently, 36,938 promoter regions were found to be meaningful in univariate Cox analysis. C-index (index of concordance, consistency index) is usually used to evaluate the performance of the model, therefore, we first calculate the C-index of each promoter region, and then select the top 5000 promoter regions ranked b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com