Methylation marker related to colorectal cancer and kit for detecting colorectal cancer
A methylation marker, colorectal cancer technology, applied in the field of biomedicine, can solve the problems of inconsistent results, limited sensitivity, low concentration, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0124] Example 1 Screening and verification of cfDNA methylation markers for colorectal cancer
[0125] 1. Colorectal cancer tissue specificity
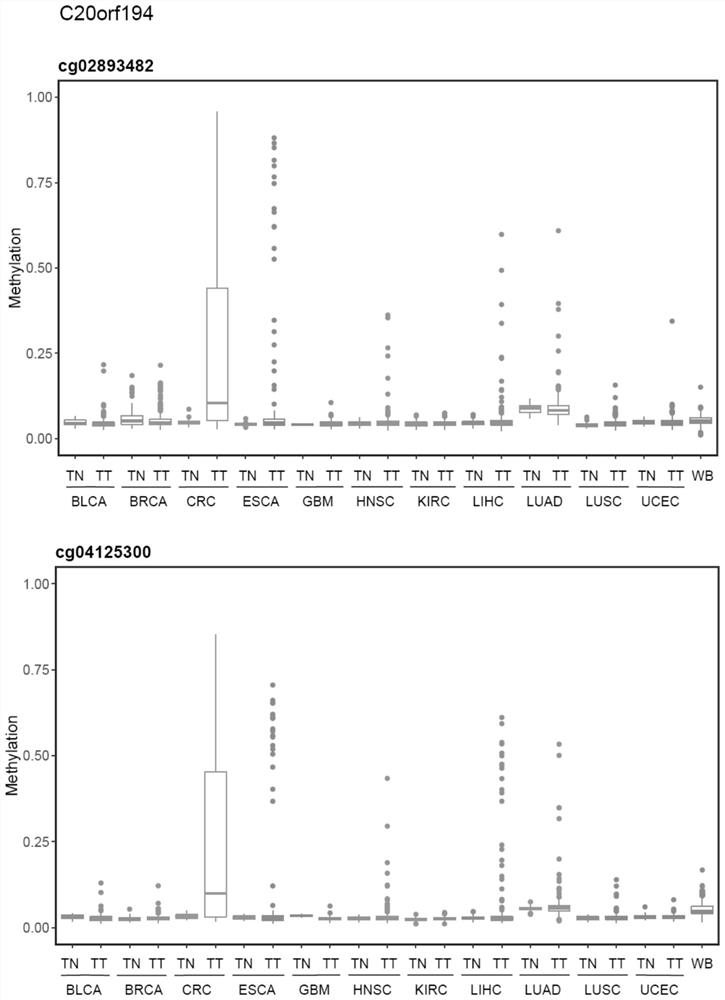
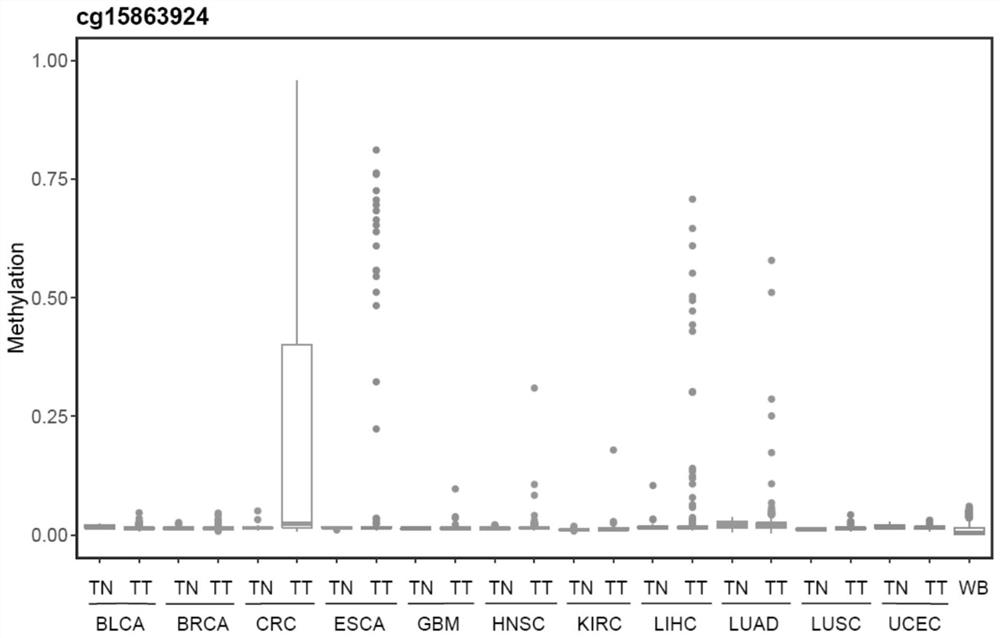
[0126] The present invention utilizes databases such as TCGA to analyze colorectal cancer tissue-specific methylation profiles, screen out tumor methylation-specific markers, and screen out 10 CpG sites mainly distributed on ZNF304, LIFR and C20orf194 genes. Based on the 450K methylation data of 11 cancer tissues including colorectal cancer in the TCGA database and healthy peripheral blood leukocytes from GEO, it shows that the 10 CpGs are specific to colorectal cancer tissues, and the average methylation level is shown in figure 1 , see Figure 2, Figure 3, and Figure 4 for the box plots of the methylation levels of the sites contained in the three genes. See Table 1 for the genome information of the 10 CpG sites, and Table 2 for the cancer types.
[0127] Table 1
[0128]
[0129]
[0130] Note: Genome annotation information...
Embodiment 2
[0176] Example 2 Methylation-specific fluorescent quantitative PCR (qMSP)
[0177] The qMSP probe method was used to detect the DNA methylation of colorectal cancer, para-cancerous tissues and peripheral blood leukocytes, and primers were designed based on the results of targeted sequencing. The target fragment plasmid was constructed using fully methylated human genomic DNA treated with bisulfite as a template, and the methylation standard curve was constructed by gradient dilution and the equation was established. The sequence without CG sites on ACTB was used as the internal reference gene, and the target gene of the sample was The ratio of the amplified copy number to the amplified copy number of the internal reference gene was used as the relative methylation level of the target gene.
[0178] 1. Primer and probe design
[0179] Design qMSP primers and probes for ZNF304, LIFR and the internal reference gene ACTB for the screened sites, the amplification primers include m...
Embodiment 3
[0209] Example 3 MethyLight ddPCR detection method and establishment of kit
[0210] The technology adopted in the present invention is digital PCR technology combined with the MethyLight method, by designing specific primers and probes for human ZNF304 and LIFR candidate methylated regions, amplifying bisulfite modified and prepared by microdropletization Measure the DNA sample, and determine whether methylation and the methylation level have occurred in the human ZNF304 and LIFR candidate methylation regions in the sample to be tested according to the ratio of the number of positive droplets amplified by PCR and the total number of positive droplets, thereby Determine whether colorectal cancer occurs in the sample to be tested.
[0211] 1. Primer and probe design
[0212] Design ddPCR primers and probes for ZNF304 and LIFR for the screened sites. Amplification primers include methylated sequence amplification primers and unmethylated sequence amplification primers. TaqMan p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com