Molecular marker for detection or curative effect evaluation of liver cancer and application
A technology of molecular markers and curative effect evaluation, applied in drug combinations, organic active ingredients, biochemical equipment and methods, etc., can solve the problems of high incidence, lack of HCC imaging features, unsatisfactory prognosis of liver cancer, etc., to achieve Excellent specificity and improved detection accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
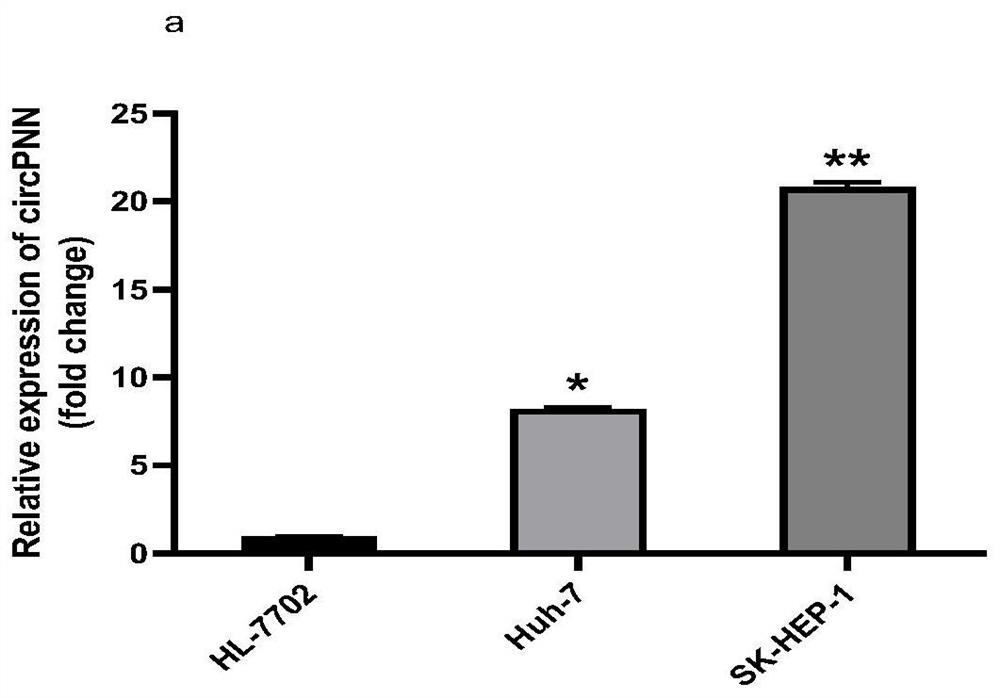
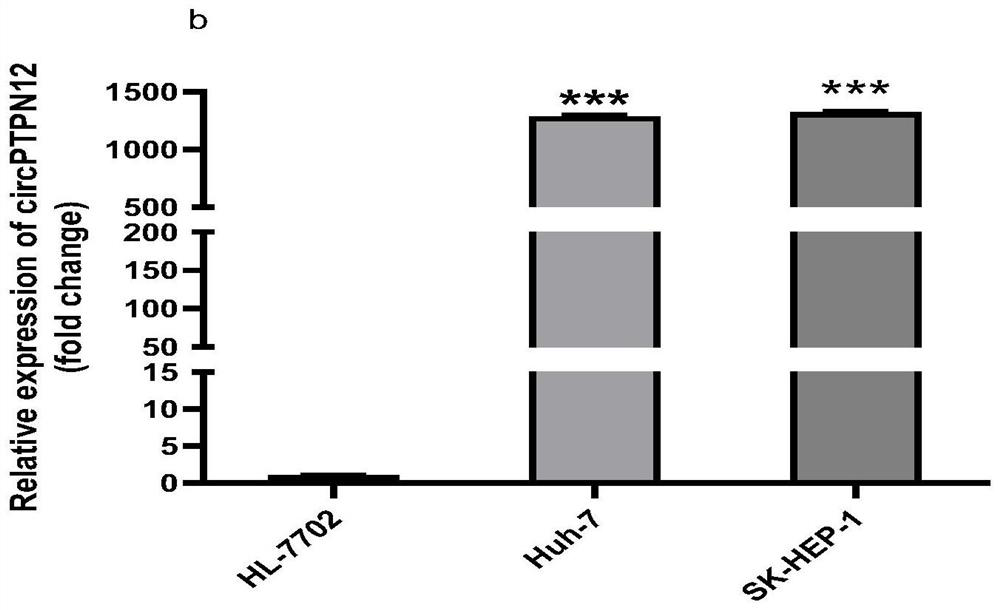
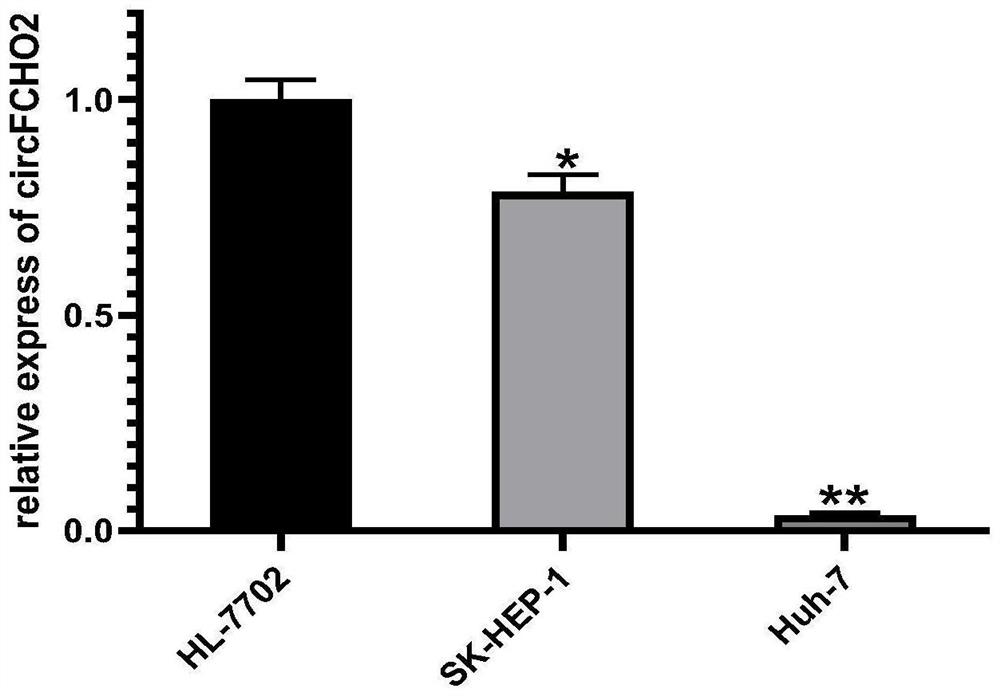
[0064] This example shows the expression of circPNN (hsa_circ_0101802), circFCHO2 (hsa_circ_0002490) and circPTPN12 (hsa_circ_0005197) in human liver cancer cell lines and human normal liver cell lines:
[0065] Human normal liver cell line HL-7702 cells use RPMI-1640 medium + 10% FBS fetal bovine serum (FBS); while Huh-7 uses DMEM medium + 10% FBS; SK-HEP-1 uses a typical culture from the Chinese Academy of Sciences Special culture medium purchased from the Cell Bank (Shanghai) of the Committee for the Preservation of Animals. The above cells were all cultured in a cell incubator at 37° C. with a CO2 saturation humidity of 5%, and grew adherently.
[0067] From a liquid nitrogen tank or a -80°C ultra-low temperature refrigerator, quickly shake back and forth in a 37°C water bath to melt the cells quickly. Use a 1ml pipette gun to transfer the cell suspension in the cryopreservation tube into a 15ml centrifuge tube, blow and mix evenly, and centrif...
Embodiment 2
[0103] This example is the effect of silencing circPNN (hsa_circ_0101802) in liver cancer cell SK-HEP-1 on cell biological behavior:
[0104] (1) Experimental grouping
[0105] ①siRNA-1 group: circPNN-siRNA-1 (sequences are SEQ ID NO:10 and SEQ ID NO:11) / lentivirus complex was added during transfection. ②siRNA-2 group: circPNN-siRNA-2 (sequences are SEQ ID NO: 12 and SEQ ID NO: 13) / lentivirus complex was added during transfection. ③Blank control group: liver cancer cells without any intervention measures.
[0106] (2) Cell transfection
[0107] Take SK-HEP-1 liver cancer cells in good growth state and logarithmic growth phase, digest with trypsin, centrifuge, resuspend in culture medium and count, adjust the cell concentration to 1.0×10 5 Inoculate into 6-well plates at the density of cells / ml and place in CO 2 Cultivate in an incubator until the cell density reaches 70%-80%; discard the old medium and wash the cells twice with PBS. Use the complete medium to dilute the 2...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com