Genetically engineered bacterium for producing L-threonine as well as construction method and application of genetically engineered bacterium
A genetically engineered bacteria and genetic engineering technology, applied in the direction of microorganism-based methods, biochemical equipment and methods, bacteria, etc., can solve the problem of the conversion rate of cofactor biotin-deficient strains, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
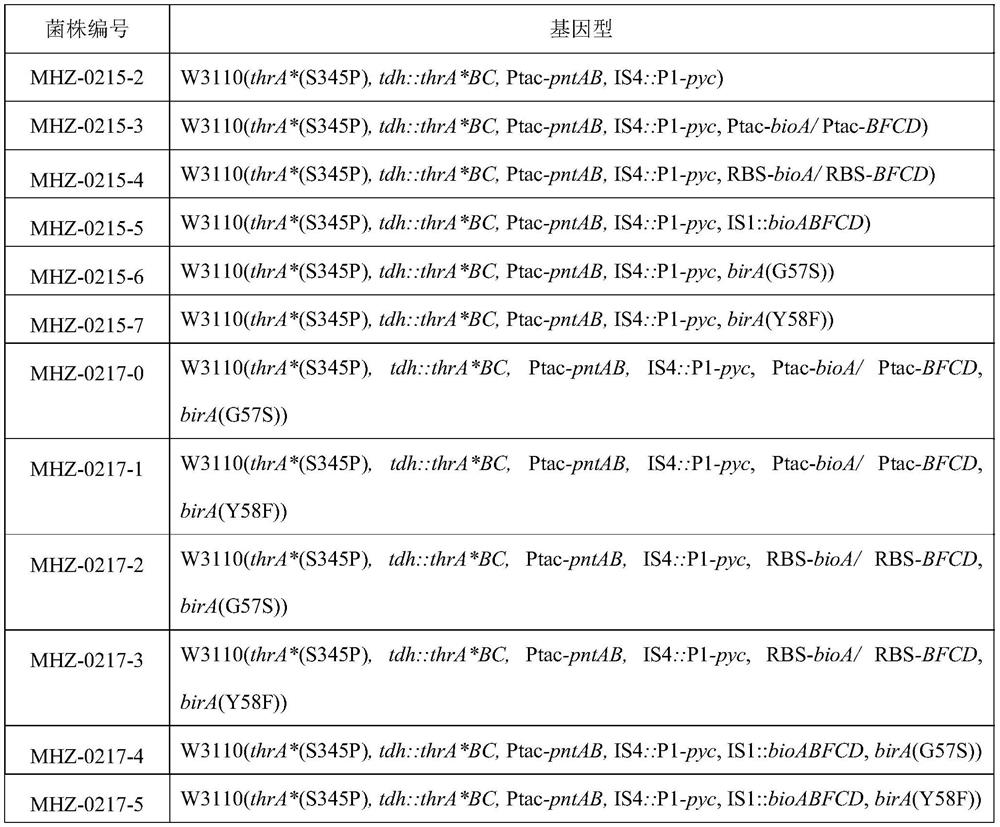
[0151] Embodiment 1 prepares the bacterial strain MHZ-0215-3 (promoter strengthening) of strengthening bioABFCD gene
[0152] (1) Construction of pTargetF-N20(tac-bioABFCD) plasmid and Donor DNA-1
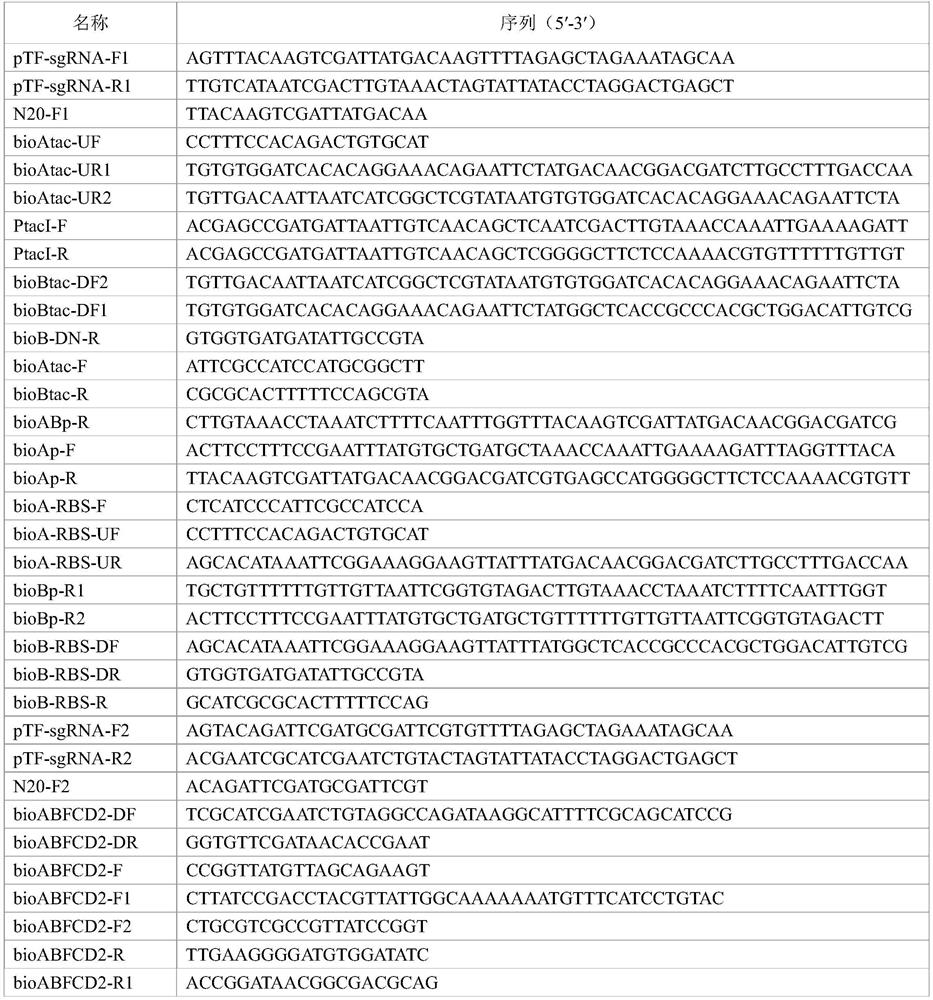
[0153] Step 1: Using the pTargetF plasmid as a template (see Multigene Editing in the Escherichiacoli Genome via the CRISPR-Cas9 System, Jiang Y, Chen B, et al.Appl.EnvironMicrobiol, 2015), select pTF-sgRNA-F1 / pTF-sgRNA - R1 primer pair, amplify the pTF linear plasmid with N20, use the seamless assembly ClonExpress kit to assemble the linear plasmid at 37°C, and then transform Trans1-T1 competent cells to obtain pTargetF-N20(tac-bioABFCD) , and carry out PCR identification and sequencing verification; Step 2: Using the W3110 genome as a template, select bioAtac-UF / bioAtac-UR1 and bioAtac-UF / bioAtac-UR2 primer pairs to amplify the upstream homology arm containing the tac promoter① ; Step 3: Using the W3110 genome as a template, select the PtacI-F / PtacI-R primer pair to amplify the ...
Embodiment 2
[0160] Example 2 Preparation of bioABFCD gene RBS enhanced strain MHZ-0215-4 (RBS-bioABFCD)
[0161] (1) Construction of pTargetF-N20 (RBS bioABFCD) plasmid and Donor DNA-2 construction
[0162] Step 1: Using the pTargetF plasmid as a template (see Multigene Editing in the Escherichiacoli Genome via the CRISPR-Cas9 System, Jiang Y, Chen B, et al.Appl.EnvironMicrobiol, 2015), select pTF-sgRNA-F1 / pTF-sgRNA - R1 primer pair to amplify the pTF linear plasmid with N20, use the seamless assembly ClonExpress kit to assemble the linear plasmid at 37°C, and then transform Trans1-T1 competent cells to obtain pTargetF-N20 (RBS bioABFCD), And carry out PCR identification and sequencing verification; Step 2: Using the W3110 genome as a template, select the bioA-RBS-UF / bioA-RBS-UR primer pair to amplify the upstream homology arm containing RBS ①; Step 3: Using the W3110 genome As a template, use bioAp-F / bioAp-R, bioAp-F / bioABp-R, bioAp-F / bioBp-R1, bioAp-F / bioBp-R2 primer pairs to amplify t...
Embodiment 3
[0169] Example 3 Preparation of the strain MHZ-0215-5 (multiple copies) of the enhanced bioABFCD gene
[0170] (1) Construction of pTargetF-N20(IS1::bioABFCD) plasmid and Donor DNA-3
[0171] Step 1: Using the pTargetF plasmid as a template (see Multigene Editing in the Escherichiacoli Genome via the CRISPR-Cas9 System, Jiang Y, Chen B, et al.Appl.EnvironMicrobiol, 2015), select pTF-sgRNA-F2 / pTF-sgRNA -R2 primer pair to amplify the pTF linear plasmid with N20, use the seamless assembly ClonExpress kit to assemble the linear plasmid at 37°C, and then transform Trans1-T1 competent cells to obtain pTargetF-N20 (IS1::bioABFCD ), and carry out PCR identification and sequencing verification; Step 2: Using the W3110 genome as a template, select the bioABFCD2-UF / bioABFCD2-UR primer pair to amplify the upstream homology arm ①; Step 3: Using the W3110 genome as a template, select bioABFCD2 -F1 / bioABFCD2-R1 primer pair to amplify the upper half of the bioABFCD operon ②; Step 4: Using th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com