Antisense RNA sequence combination and method for inhibiting gonad development of pelteobagrus fulvidraco and application
An antisense technology for yellow catfish, which is applied in the fields of molecular biology and breeding biology, can solve the problems of high consumption, difficulty in growing seedlings to market specifications, prolonging the marketing cycle of yellow catfish, etc., achieving high success rate and shortening the marketing time Cyclic, representative and stable effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] An antisense RNA sequence combination for inhibiting the gonad development of pelagic catfish consists of Anti-FSHR-1 and Anti-FSHR-2, wherein the nucleotide sequence of Anti-FSHR-1 is:
[0051] 5'-AAAAACATGTCCCCTGCGTGCATCATGAGCCAAGAGAGTGTGAAGCGCAGCATAGCACCCAGACCATCTTCTTCATTTTTATGAAGAAATGCCCTCCTA-3' (SEQ ID No. 1).
[0052] The nucleotide sequence of Anti-FSHR-2 is:
[0053] 5'- AGATCATATGTAGACGACAATAGCTTATGGTGTATGAATTTGGAAAGTTTTTGGGGGTCAGTGGGGATGAGCAACATGTCCCAGTGAGTATAGCGTCCCA-3' (SEQ ID No. 2).
Embodiment 2
[0055] A method for inhibiting the development of the gonads of the peliotus catfish, the steps are as follows:
[0056] 1) After the two antisense RNA sequences in Example 1 were synthesized, they were cloned into the pcDNA3.1 expression vector between the XhoI and XbaI sites (containing the highly expressed CMV promoter), and this product was used as a subsequent PCR amplification template.
[0057] 2) PCR amplification: design a pair of specific primers for template amplification, the specific primers are PolyAF2 and PolyAR1; wherein, the nucleotide sequence of polyAF2 is 5'-TTTTGCGCTGCTTCGCGATGTAC-3' (SEQ ID No.3); The nucleotide sequence of the primer polyAR1 is 5'-TCCCAATCCTCCCCCTTGCTG-3' (SEQ ID No.4). The reaction system is 50 μL and consists of the following components: 25 μL of 2×Mastermix, 5 μL of primers, 18 μL of ultrapure water, and 2 μL of template. The amplification program was: pre-denaturation at 95°C for 2 min, 34 cycles (denaturation at 95°C for 30 s, ann...
Embodiment 3
[0065] Test plan: the newly hatched larvae of Example 2 were placed in a small-scale circulating water system and raised for 60 days, and then transferred to a large-scale circulating water system for breeding. The breeding cycle is 12 months. Each test group was fed extruded feed twice a day, and the feeding amount was 5% to 15% of body weight. The feed contains 40% protein and 8% fat.
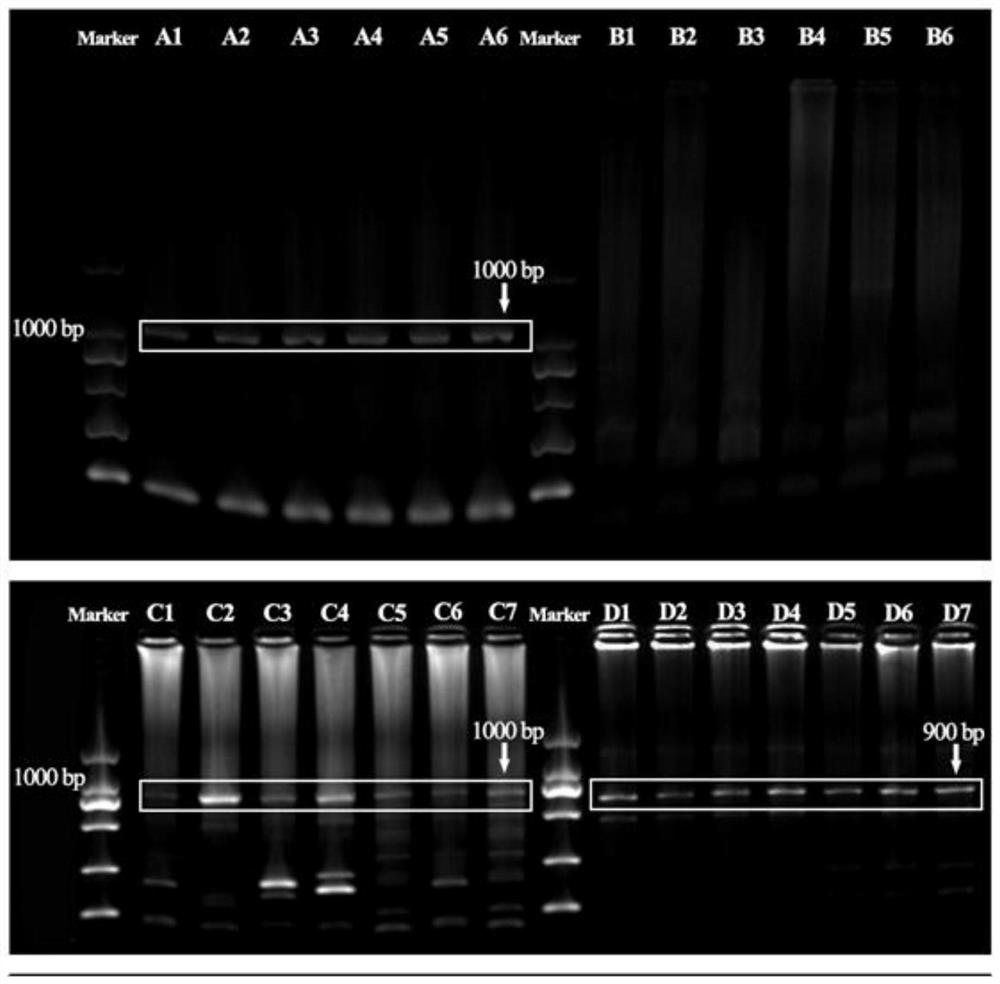
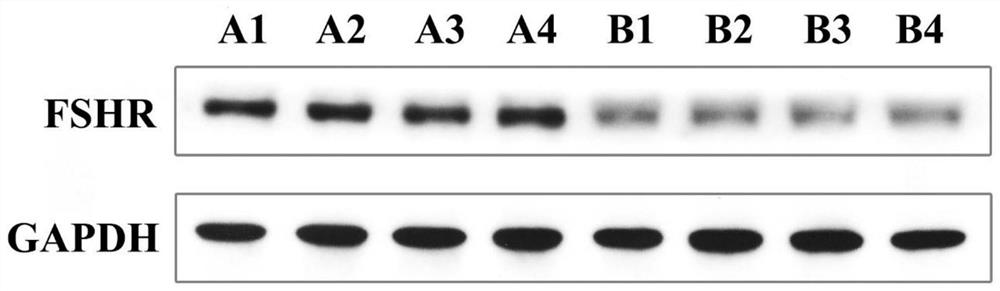
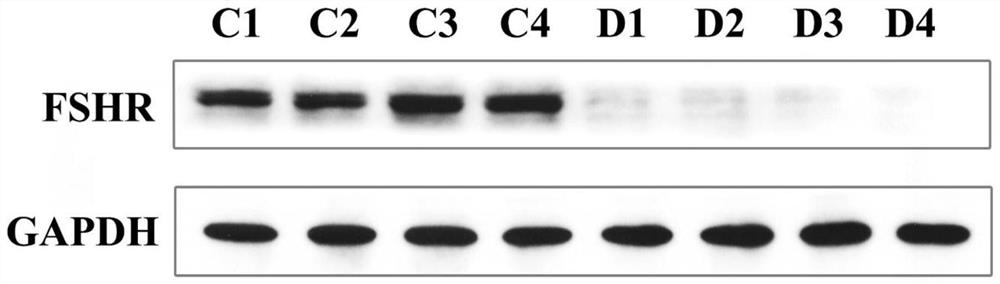
[0066] PCR and agarose gel electrophoresis to verify transfection effect
[0067] After 200 days of breeding, randomly select 13 test fish of the antisense RNA fragment transfection group (test group), 7 test fish of the negative control group (negative control group) and 6 test fish of the control group (control group) to cut off the gonads respectively. Tissues were tested for transfection effect, and genomic DNA was extracted using TakaRa MiniBEST Universal Genomic DNAExtraction KitVer 5.0 kit. 20 μL reaction system, including: upstream and downstream primers (polyAF2: TTTTGCGCTGCTTCGCG...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com