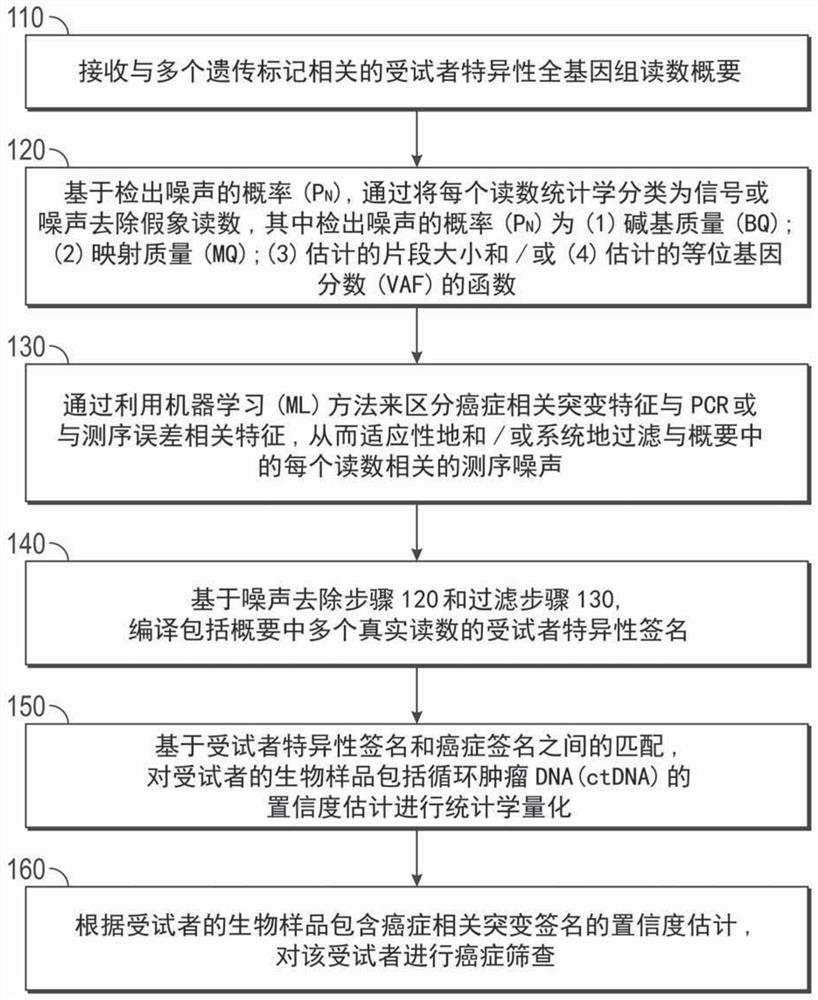
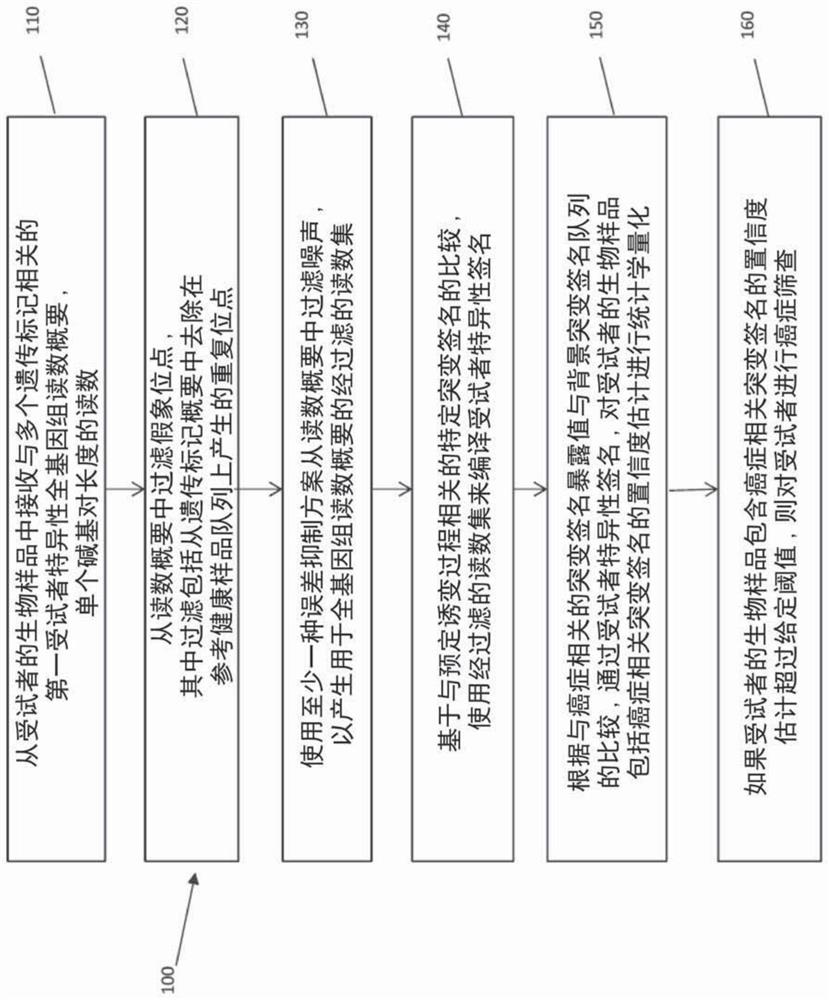
Ultra-sensitive detection of circulating tumor DNA through genome-wide integration
A genome-wide, read-out technology, applied in the field of medical diagnosis, which can solve problems such as poor imaging technology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0373] Example 1: Design of a Somatic Mutation Classifier
[0374]When designing models for classification of somatic mutations, it is important to identify sources of error that can lead to false positive somatic mutations. True mutations are likely to be of high base quality regardless of position in the read. Likewise, the read base, reference base, and alignment string (CIGAR) at the position of the true mutation may not be relevant to the read alignment. More specifically, true somatic mutations can be expected to be spatially invariant. It is well known that systematic errors in sequencing experiments depend on position in the reads, so while a mutation itself may be spatially invariant, its position in the reads is often not. Errors caused by mismapping are likely to contain repetitive sequences or very specific sequence motifs (eg TTAGGG in telomeres). Therefore, models that accurately represent the spatial invariance in real somatic mutations and errors due to mapp...
Embodiment approach
[0424] Based on the foregoing, the system and method can be developed as a complete early detection engine. Although the engine captures position in a read by using a fully connected sigmoid layer, some architectures may be better suited for capturing relative position in a read. Furthermore, additional sources of information contained in read pairs from DNA fragments, which were not included in the primary detection, can be used to determine the strand of origin (Watson or Crick) and estimate the size of the DNA fragment. It has been observed that ctDNA has a different fragment size distribution compared to regular circulating healthy DNA (Underhill et al., PLoS Genetics, 12(7):e1006162, 2016).
[0425] The aforementioned systems and methods can be integrated with a recurrent neural network (RNN). RNNs have now been shown to be a powerful tool for using length as a feature in bioinformatics at distances even up to 1kb, well beyond the size of ctDNA fragments (Hill et al., bi...
Embodiment approach 2
[0426] Embodiment 2: Methods and systems for detecting and validating tumor-specific low-abundance tumor markers and their use in cancer diagnosis
[0427] The disclosed systems and methods can be used for early diagnosis of cancer. As is known in the art, the abundance of ctDNA limits targeted sequencing techniques in the case of early stage cancer or residual disease detection compared to metastatic cancer (characterized by high disease burden and significantly elevated ctDNA) usage of. Considering the known limited amount of cfDNA in the setting of low tumor burden, first, the potential to optimize cfDNA extraction was investigated. First, to reduce variance derived from sample collection and inter-individual variation, uniform cfDNA material generated from large-volume plasma collection (approximately 300 cc) by plasmapheresis from healthy subjects and cancer patients undergoing hematopoietic stem cell collection was used, Commercially available extraction kits and metho...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com