Method and device for detecting chromosome joint deletion and storage medium
A chromosome and mutation detection technology, applied in the field of chromosome detection, can solve the problems of 1p/19q chromosome joint deletion, etc., and achieve the effect of improving the use efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
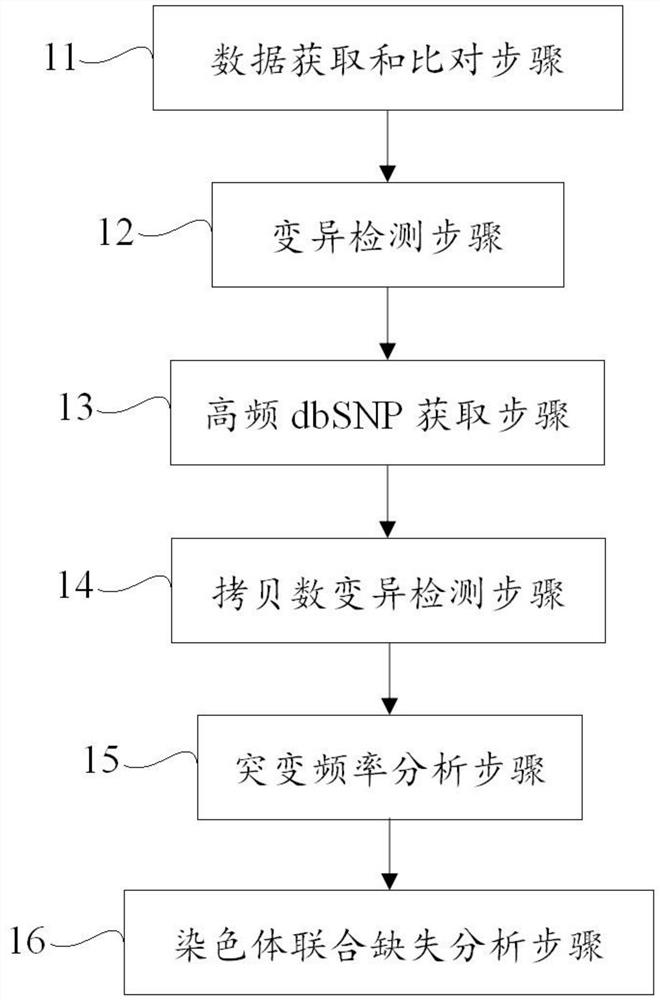
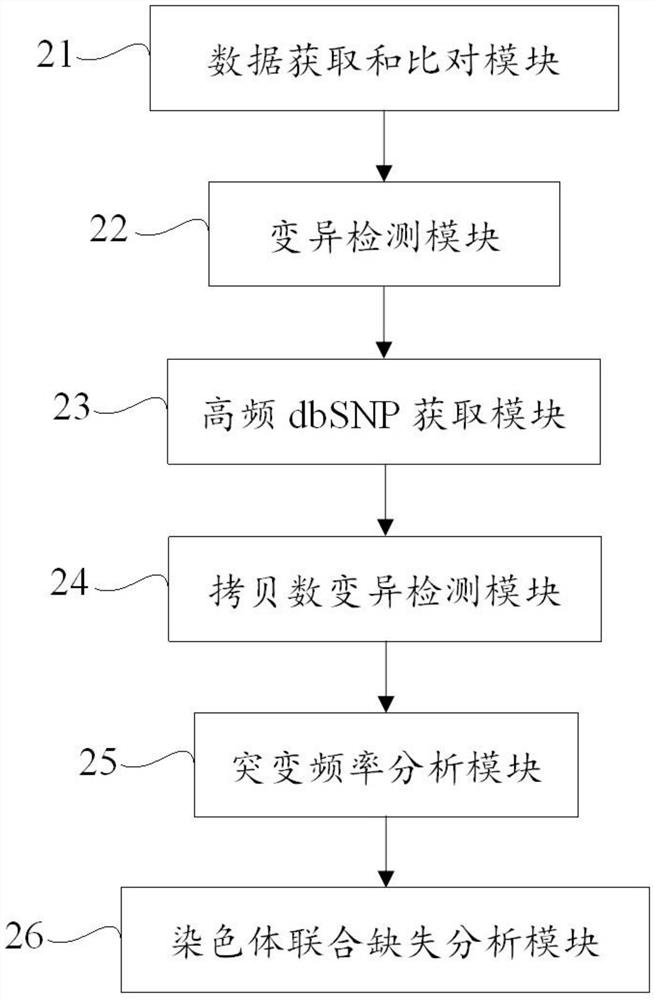
[0070] In this example, the capture probes targeting the 1p and 19q regions are used to hybridize and capture genomic DNA, and then perform high-throughput sequencing; this example analyzes and detects chromosome loss of heterozygosity based on high-throughput capture sequencing. Specifically, this example Loss of heterozygosity on chromosome 1p and 19q was analyzed by combining point mutation frequency and copy number variation. Specific steps include:
[0071] The capture sequencing results of the short arm of chromosome 1 and the long arm of chromosome 19 of the tumor tissue sample and the corresponding normal control sample were respectively obtained, and the capture sequencing results of the tumor tissue sample and the normal control sample were compared to the reference genome and deduplication was obtained. The data after deduplication, and then perform the following operations:
[0072] a, Read the SNP result file generated by VarScan2 to obtain the variation informat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com