Method for efficiently expressing acid protease and application thereof
A technology of acid protease and fusion protein, which is applied in the field of high-efficiency expression of acid protease, and can solve the problems of low activity of acid protease, limited application of acid protease, reduction of catalytic efficiency, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] The construction of embodiment 1 recombinant Pichia pastoris
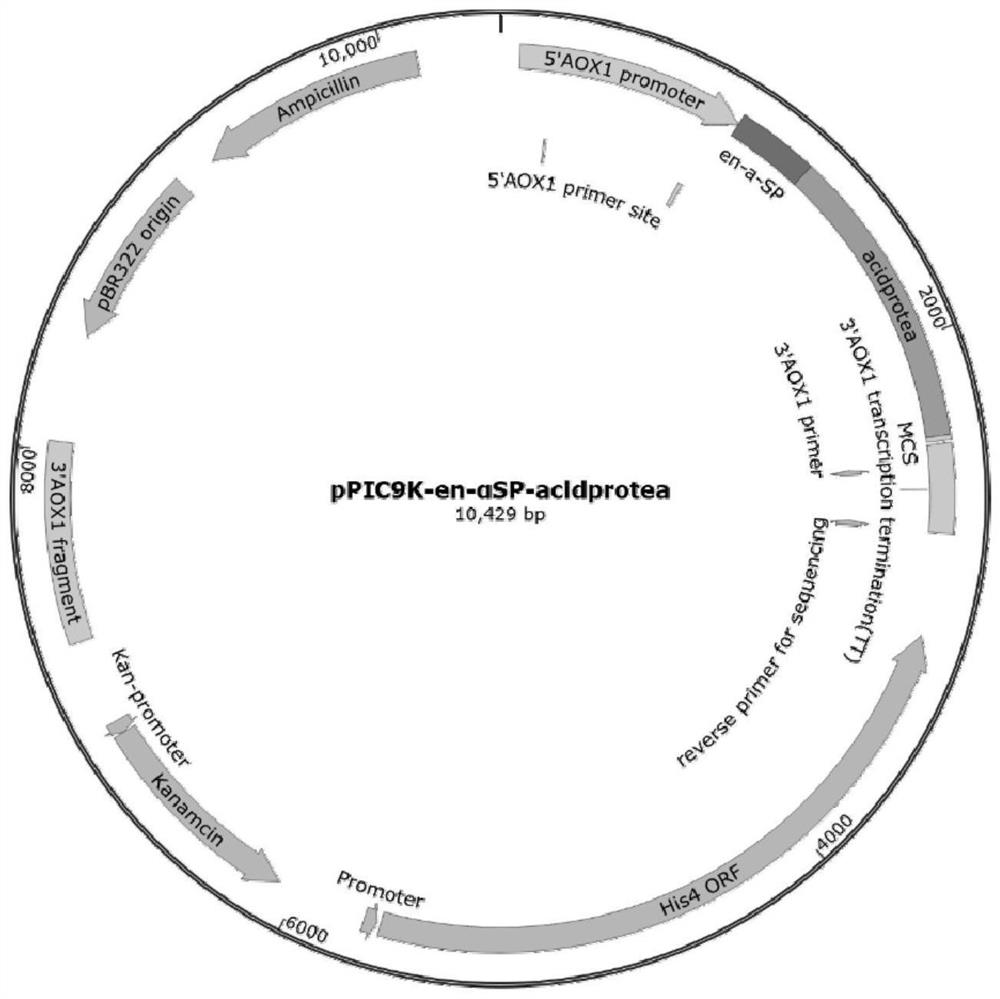
[0035] (1) Construction of recombinant plasmid pPIC9K-en-αSP-acidprotea
[0036] The enhanced α-signal peptide (en-αSP) whose nucleotide sequence is shown in SEQ ID NO.4 is connected to the N-terminus of the acid protease shown in SEQ ID NO.2, and artificially synthesized, Add Bam HI and Not I restriction sites at the beginning and the end respectively, and integrate the sequence connected with the restriction sites into the cloning vector pUC57 to form the plasmid pUC57-enα-SP-acidprotea. The plasmid pUC57-enα-SP-acidprotea was double-digested with Bam HI and Not I to obtain a 1428bp sequence containing enhanced α-signal peptide and acid protease and recovered, and then the vector pPIC9K was obtained by double-digestion with Bam HI and Not I The 8998bp carrier sequence was recovered, and the sequence containing the enhanced α-signal peptide and acid protease was connected with the carrier sequence with DNA...
Embodiment 2
[0042] Embodiment 2 Recombinant Pichia pastoris shakes flask fermentation to produce acid protease
[0043] From the plate containing recombinant Pichia pastoris GS115 / pPIC9K-en-αSP-acidprotea obtained in Example 1, pick 10 single colonies and inoculate them in 25mL liquid medium YPD, and cultivate them at 30°C and 220rpm 24h, room temperature 6000rpm, centrifuge 5min, collect the cells, discard the supernatant, resuspend the cells with 30mL liquid medium YP, induce expression at 28°C, 220rpm, add methanol to the final concentration of 1% every 24h to continue the induction, shake After 72 hours of bottle induction, the flask was shaken to obtain a fermentation broth.
[0044] The recombinant strain P. pastoris GS115 / pPIC9K-acidprotea obtained in Example 1 containing the original α-signal peptide was used as a control strain, and the induction culture was carried out at the same time.
[0045] When the final flask was shaken, the enzyme activity of the control strain using th...
Embodiment 3
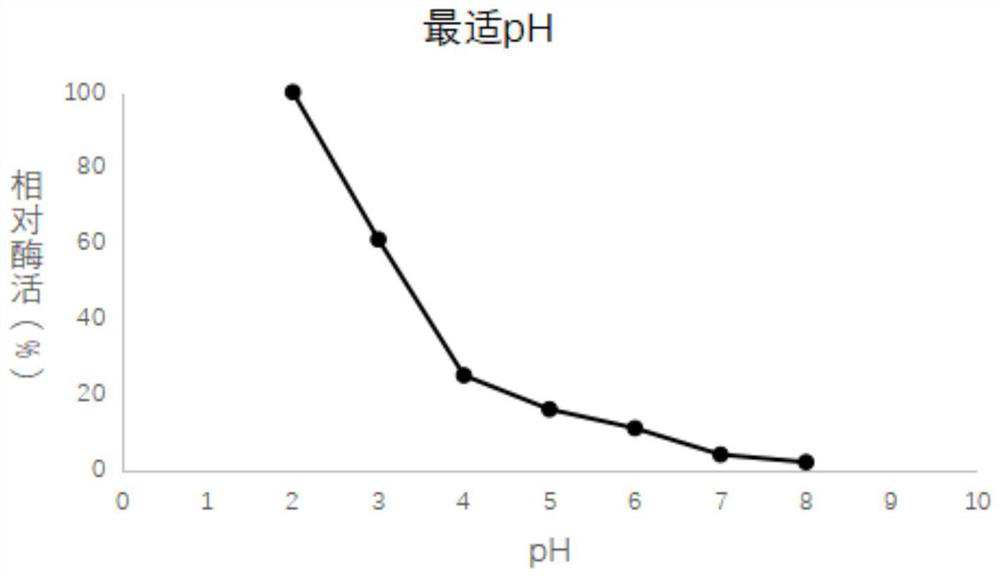
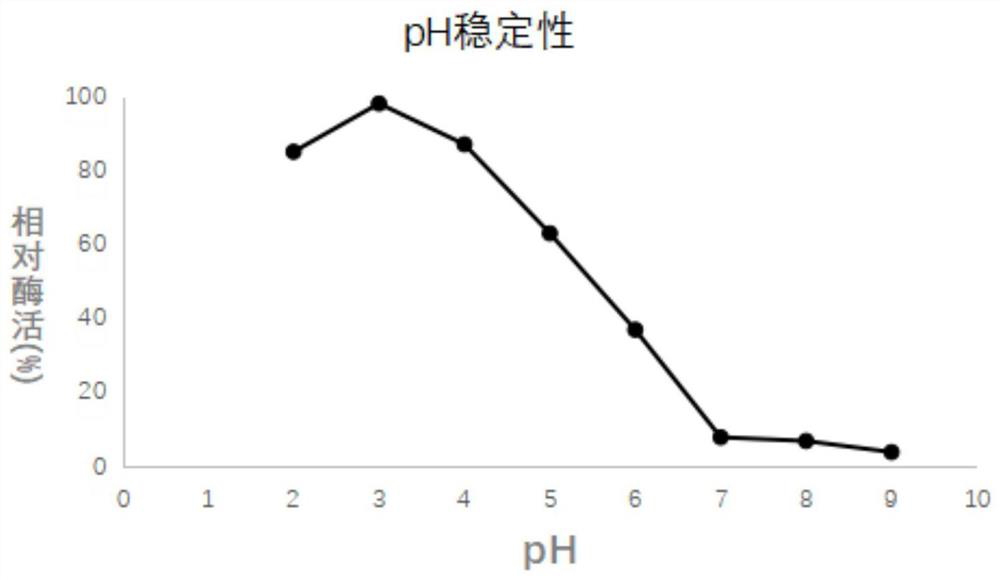
[0046] The enzymatic performance of embodiment 3 recombinant acid proteases
[0047] (1) Purification of acid protease
[0048] The fermentation broth obtained in Example 2 was centrifuged at 4°C at 8000r / min for 10min, and the fermentation supernatant was collected to obtain a crude enzyme solution, using 20mmol / L NaH 2 PO 4 -Na 2 HPO 4 (p H 6.5) buffer solution and 10kDa ultrafiltration membrane bag were used to dialyze the crude enzyme solution. The dialyzed crude enzyme solution was collected, centrifuged at 4°C and 12000r / min for 30min, and the supernatant was collected.
[0049] The protein was purified using HitrapTM Q FP (5mL) anion exchange chromatography column. Use the AKTA protein purifier to separate and purify the protein of the crude enzyme solution. The column purification conditions are as follows: use 10 times the column volume of buffer A (20mmol / LNaH2PO4-Na2HPO4 buffer solution, pH 6.5) to equilibrate the anion column, and use 1mL / min Injection at a f...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com