Method for one-step transformation and marking of blue-green algae chromosomes and application
A marking method and chromosome technology, which is applied in the field of fluorescent labeling of cyanobacterial chromosomes, can solve the problems of limited research on cyanobacterial chromosome replication and separation, lack of effective labeling of cyanobacterial chromosomes, and the number of chromosomes is not fixed.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1, Construction of Chromosomal Marker Vector 277ARY
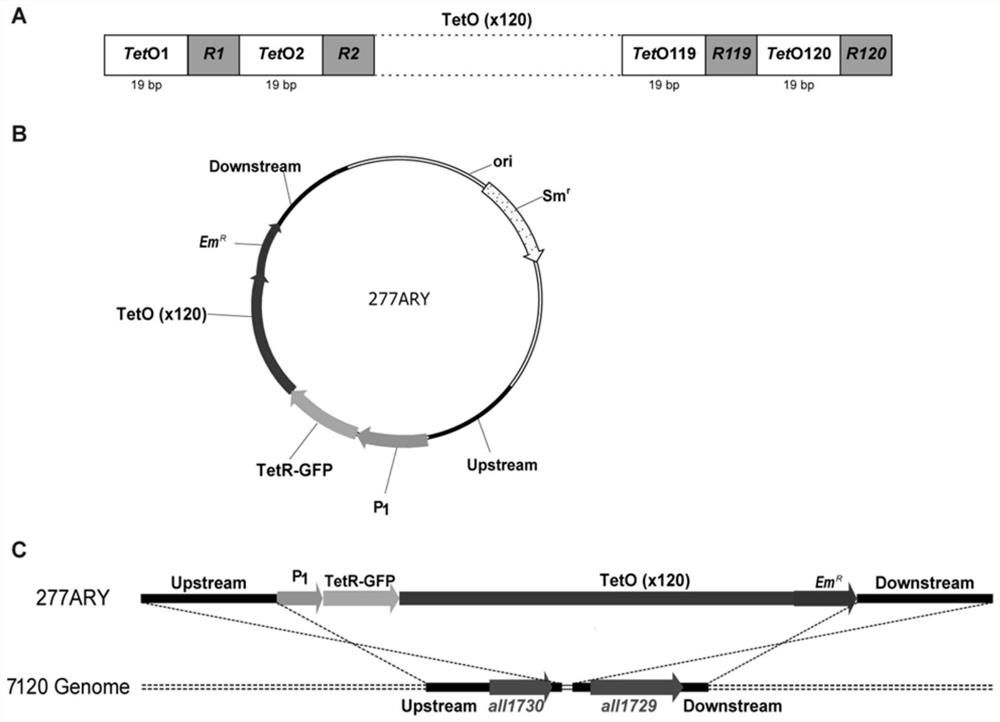
[0030] (1) Construction of TetO repeat sequence TetO×120
[0031] The schematic diagram of constructing the TetO sequence array TetO×120 formed by repeating the TetO sequence 120 times is as follows figure 1As shown in A, wherein the base sequence of TetO is tctctatcactgataggga. Because a continuous repeated sequence is very unstable in the genetic replication of bacteria, a random base sequence R of 10-30 nt is inserted between every two TetO. In a case we implemented, the entire nucleotide sequence of the designed TetO×120 is shown in SEQ ID No:2 in the sequence listing. Then TetO×120 was obtained by Beijing Qingke Biotechnology Co., Ltd. through gene synthesis technology.
[0032] (2) Construction of chromosome marker vector 277ARY
[0033] The schematic diagram of the construction of the chromosome marker vector 277ARY is shown in figure 1 As shown in B, this carrier has the core element Upstream—P ...
Embodiment 2
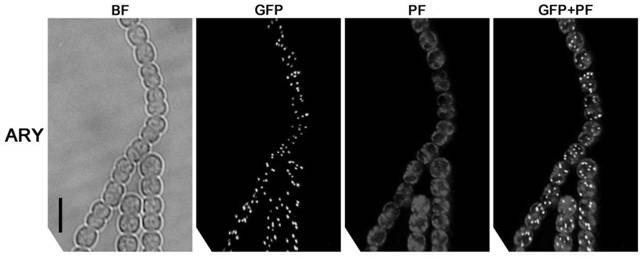
[0034] Example 2, Screening of Chromosomal Marker Positive Strain ARY and Chromosomal Fluorescent Detection
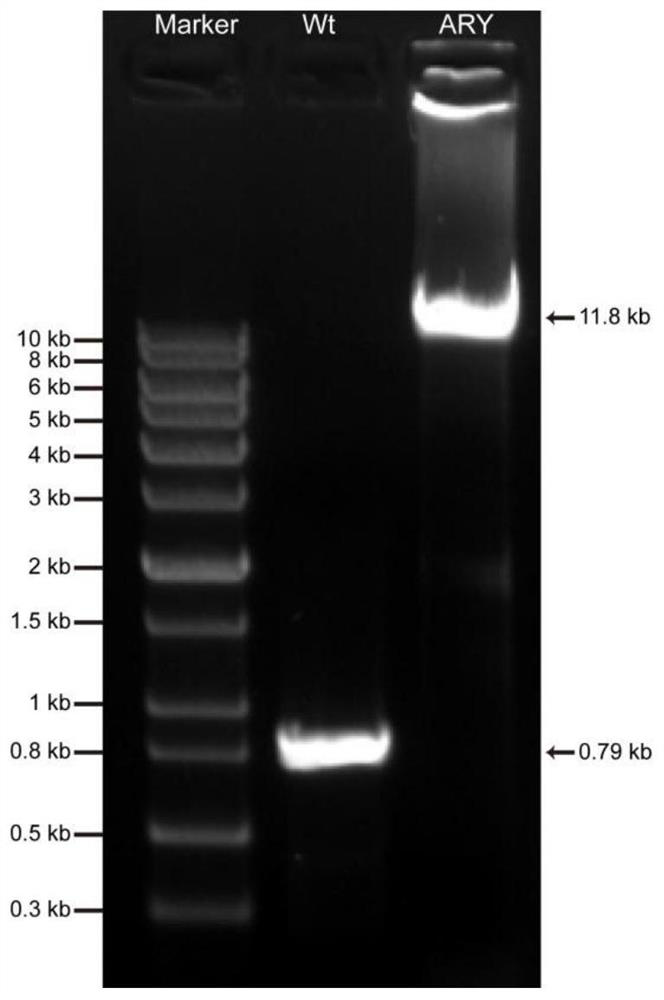
[0035] (1) Screening and homozygosity verification of chromosomal marker-positive strain ARY
[0036] The constructed chromosomal marker carrier 277ARY was transferred into the genome of wild-type Anabaena sp. figure 1 In C, the chromosomal marker vector 277ARY in P 1 The —TetR-GFP—TetO×120—Em fragment was exchanged between the all1730 and all1729 genes of the genome chromosome of Anabaena sp.PCC 7120. The method of three-parent mating was as follows: 1. Combining the plasmid pRL443 and the helper plasmid pRL528 into Escherichia coli HB101, Recombinant strain A containing resistance genes Amp (ampicillin), Sm (streptomycin), Cm (chloramphenicol) and Tc (tetracycline) was obtained. 2. Transfer the chromosomal marker carrier 277ARY into the recombinant bacteria A to obtain the recombinant bacteria B, and wash with LB medium for 3 times. 3. Wash wild-type Anabaena sp. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com