Enzyme activity determination method of DNA ligase
A technology of DNA ligase and assay method, applied in the field of biochemistry, can solve problems such as low sensitivity, complicated operation, and inaccurate quantitative analysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] Embodiment 1, PBCV-1 DNA ligase / SplintR @ Ligase activity assay
[0060] 1. Sequence design
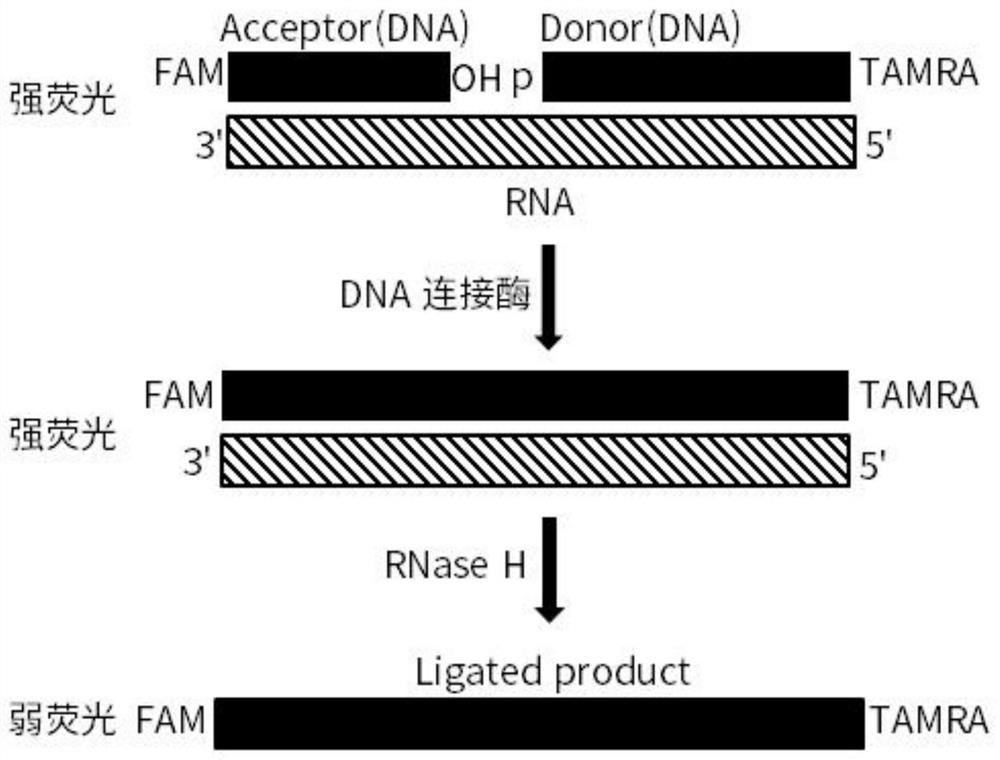
[0061] Acceptor DNA sequence:
[0062] 5'- FAM TATGACTCTACTACTAATGT- OH -3' (SEQ ID NO: 1);
[0063] Donor DNA sequence:
[0064] 5'-pAGATGCGACCTACAATGTACTAGAAGTGTC-3' TAMRA (SEQ ID NO: 2);
[0065] Paired RNA-seq:
[0066] 5'-GACACUUCUAGUACAUUGUAGGUCGCAUCUACAUUAGUAGUA GAGUCAUA-3' (SEQ ID NO: 3);
[0067] 2. Annealing of DNA / RNA hybrid duplex
[0068] (1) Prepare the DNA and RNA single strands to be annealed to 80 μM with DEPC water.
[0069] (2) DNA / RNA hybrid double-strand annealing reaction system is shown in Table 1:
[0070] Table 1
[0071]
[0072]
[0073] (3) The PCR instrument is set as follows to carry out the annealing reaction:
[0074] 1. 90°C (1min) denaturation; 2.90°C, 5s, -0.1°C each cycle; 3. GO TO Step2 650cycles) annealing; 4°C storage.
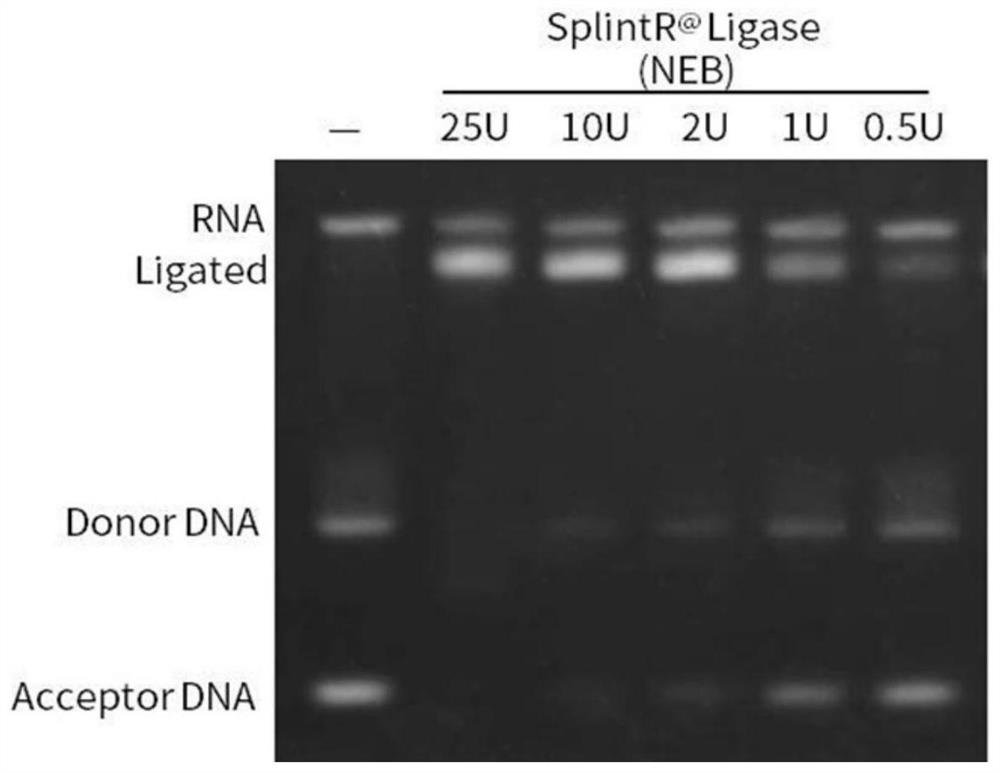
[0075] 3. SplintR @ ligase ligation reaction
[0076] SplintR used @ The ligase is NEB (25U / μl, M...
Embodiment 2
[0106] The enzyme activity assay of embodiment 2, T4 DNA ligase
[0107] The sequence to be connected is the same as the DNA / RNA hybrid strand obtained after annealing in Example 1.
[0108] 1. T4 DNA ligase ligation reaction
[0109] (1) Connection reaction system
[0110] The T4 DNA ligase used was commercial Beyotime T4 DNA ligase (1000 U / μl, D7006). The mother solution of Beyotime T4 DNA ligase 1000U / μl was diluted with its storage solution (20mM Tris, pH 7.5, 50mM KCl, 1mMDTT, 0.1mM EDTA, 50% (v / v) glycerol), and diluted to 500U / μl respectively, 250U / μl, 125U / μl, 62.5U / μl, 31.25mU / μl, 15.6U / μl, respectively add 4μl, 3μl, 2.5μl of T4 DNA ligase 1000U / μl mother solution to the reaction system according to Table 6 And 2.5 μl of a series of diluted enzyme solutions for experiments.
[0111] Table 6
[0112]
[0113] Ligation reaction temperature: 37°C, incubate for 4h.
[0114] Termination reaction: add 10 U of Thermostable RNase H (NEB M0523S) to each sample, incuba...
Embodiment 3
[0124] Example 3, Enzyme Activity Detection Based on Other Sequences
[0125] The present inventors also designed sequences different from the Acceptor DNA, Donor DNA and paired RNA in Example 1, and carried out the ligation reaction.
[0126] 1. Sequence Design 2
[0127] Acceptor DNA sequence (SEQ ID NO: 4):
[0128] 5'- FAM AGATGGATGCTCAGACTACAGT- OH -3';
[0129] Donor DNA sequence (SEQ ID NO:5):
[0130] 5'-p ATTATATGATTGGTACTCGGTCTCGCTAGGCA-3' TAMRA ;
[0131] Paired RNA sequence (SEQ ID NO:6):
[0132] 5'-UGCCUAGCGAGACCGAGUACCAAUCAUAUAAUACUGUAGUCUGAGCAUCCAUCU-3';
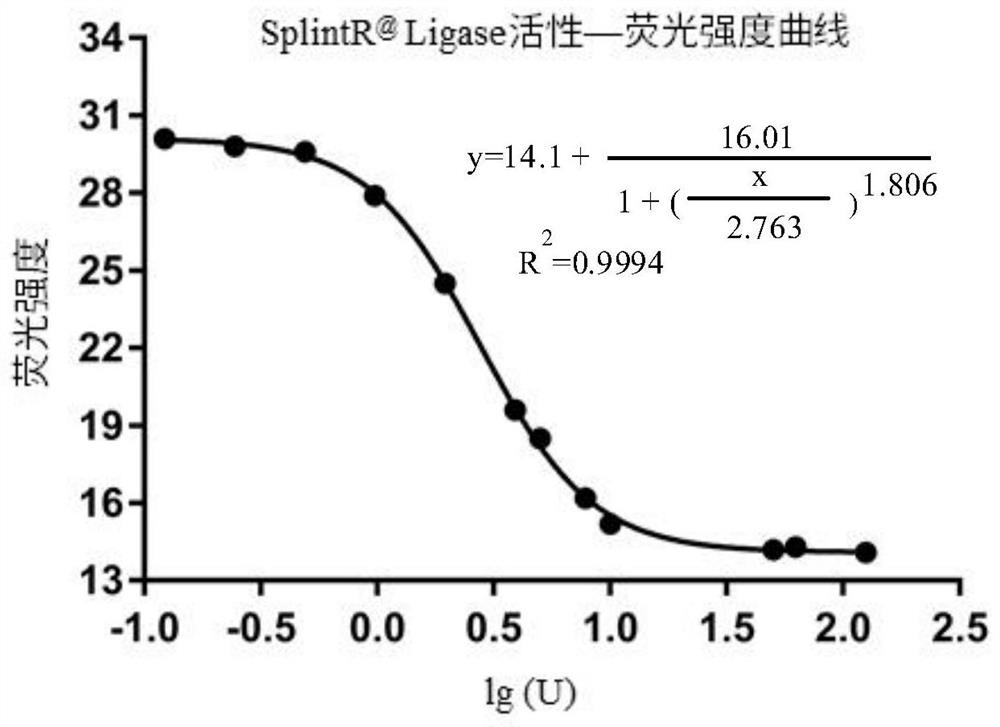
[0133] Like methods 2-5 in Example 1, the inventors used these sequences to detect the enzyme activity of SplintR@ligase, the fluorescence value changed sensitively, and the enzyme activity results were accurate. From fluorescence detection to data processing, the required time is only about 20 minutes.
[0134] 2. Sequence design 3
[0135] Acceptor DNA sequence (SEQ ID NO: 7):
[0136] 5'- FAM ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com