Method for predicting RNA binding protein based on circRNA position information
A position information and protein-binding technology, applied in the field of genetic engineering, can solve constraints and other problems, and achieve the effect of convenient and fast operation and clear process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
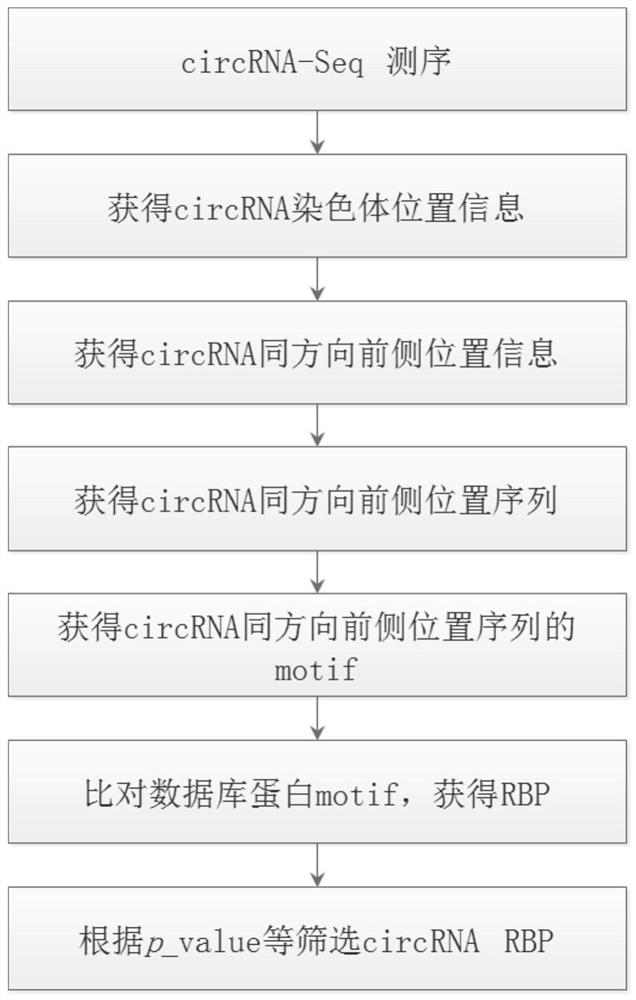
[0052] see figure 1 , the present embodiment provides a method for predicting RNA-binding proteins based on circRNA position information, comprising the following steps:
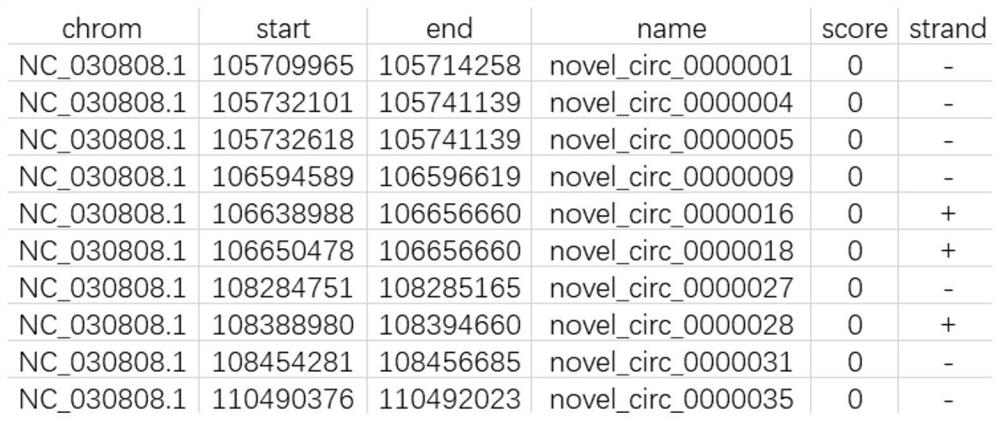
[0053] (1) Extract total RNA from samples such as tissues, cells, or blood, perform high-throughput circRNA sequencing, and obtain circRNA chromosome location information. Based on the circRNA chromosome location information, organize all circRNA chromosome location information into bed files with the suffix .bed ( Such as figure 2 shown), named circRNA_location.bed. The file has 6 columns, which are chrom: the chromosome to which the circRNA belongs, start: the coordinates of the starting position of the circRNA, end: the coordinates of the end position of the circRNA, name: the ID obtained by circRNA sequencing, score: the default is 0, strand: the direction of the circRNA on the chromosome , where + represents positive chain, - represents reverse chain;
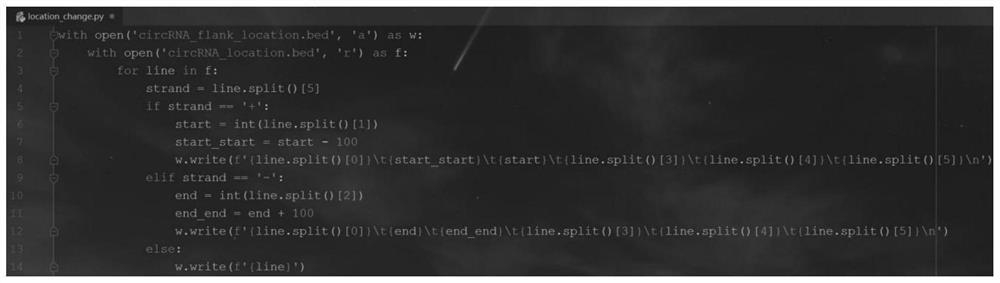
[0054] (2) Based on the circRNA_location.bed f...
Embodiment 2
[0072] This embodiment provides a method for predicting RNA-binding proteins based on circRNA position information, taking Sichuan black goat as an example, including the following steps:
[0073] (1) Extract total RNA from the endometrium sample of black goat in central Sichuan, perform high-throughput circRNA sequencing, and obtain circRNA chromosome position information, and other operations are the same as step (1) in Example 1;
[0074] (2) with embodiment 1 step (2);
[0075] (3) Download the goat reference genome nucleotide sequence file (GCF_001704415.1_ARS1_genomic.fna) from NCBI, and other operations are the same as step (3) in Example 1;
[0076] (4) The specific operation is the same as step (4) of Example 1, wherein 15 motifs are obtained through p-value screening;
[0077] (5) Download the meme format file JASPAR2020_CORE_Vertebrates_non-redundant_pfms.meme of the motifs of known proteins from the JASPAR CORE database, and other operations are the same as step (...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com