Cytosine deaminase and cytosine editor comprising same
A technology of cytosine deaminase and cytosine, which is applied in the direction of enzymes, hydrolases, chemical instruments and methods, and can solve problems affecting the efficiency of cytosine base editing tools and the application of cytosine base editing tools
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Example 1. Construction of cytosine editor in vitro expression vector and purification of cytosine editor in vitro expression
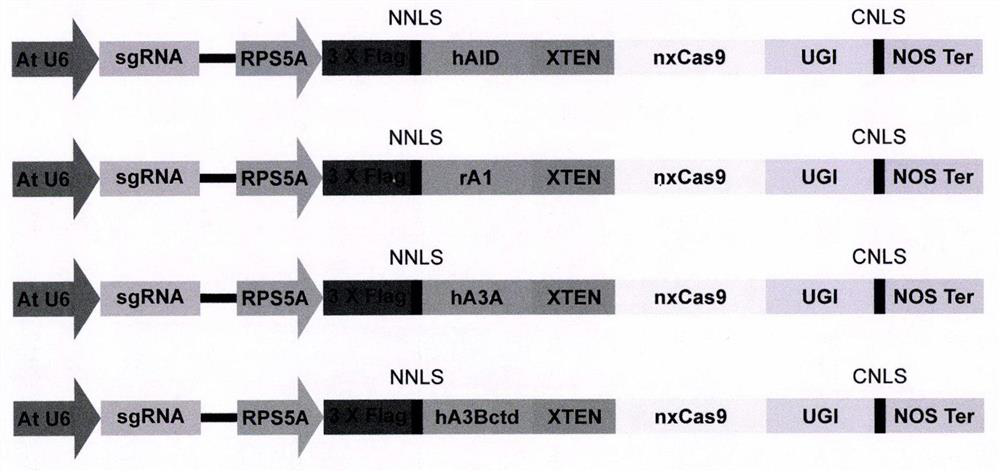
[0036] First, insert the coding sequence of MSB tag protein (SEQ ID NO.: 7) between the NcoI and BamHI restriction sites of the prokaryotic expression vector pET28a, the purpose is to enhance the expression of the prokaryotic protein in Escherichia coli, and then between BamHI and SalI respectively The coding sequences for AID-CBE, A1-CBE, A3A-CBE, A3B-CBE and A3Bctd-CBE (SEQ ID Nos.: 8, 9, 10, 11 and 12, respectively) were inserted. Among them, AID-CBE represents the fusion protein of human AID-XTEN-nxCas9-UGI, A1-CBE represents the fusion protein of rat APOBEC1-XTEN-nxCas9-UGI, and A3A-CBE represents the fusion protein of human APOBEC3A-XTEN-nxCas9-UGI , A3B-CBE indicates the fusion protein of human APOBEC3B-XTEN-nxCas9-UGI, and A3Bctd-CBE indicates the fusion protein of the C-terminal domain of human APOBEC3B-XTEN-nxCas9-UGI, both of which...
Embodiment 2
[0038] Example 2. Preparation of guide RNA
[0039] Between the HindIII and EcoRI restriction sites of plasmid pUC19, the insertion sequence is SEQ ID NO.: 13, the 20 bases at the 5' end are the T7 promoter sequence, and the 76 bases at the 3' end are the scaffold structure of the guide RNA sequence.
[0040] Using the T7 promoter and target-specific forward primers in Table 1 and the general reverse primers specific to the guide RNA scaffold structure, the three sgRNA in vitro transcription templates were obtained by PCR amplification from the above-mentioned recombinant pUC19 vector, that is, each amplification An additional primer pair is used to amplify an sgRNA template for in vitro transcription. The specific PCR system is: 10×KOD-Plus-Neo buffer, 5 μl; 2mM dNTP, 5 μl; 25mM MgSO 4 , 3μl; 10pM / μl forward and reverse primers, 1.5μl each; template plasmid, 10ng; KOD-Plus-Neo (1.0U / μl), 1μl; water up to 50μl. The PCR program is: 94 degrees for 2 minutes; 98 degrees for ...
Embodiment 3
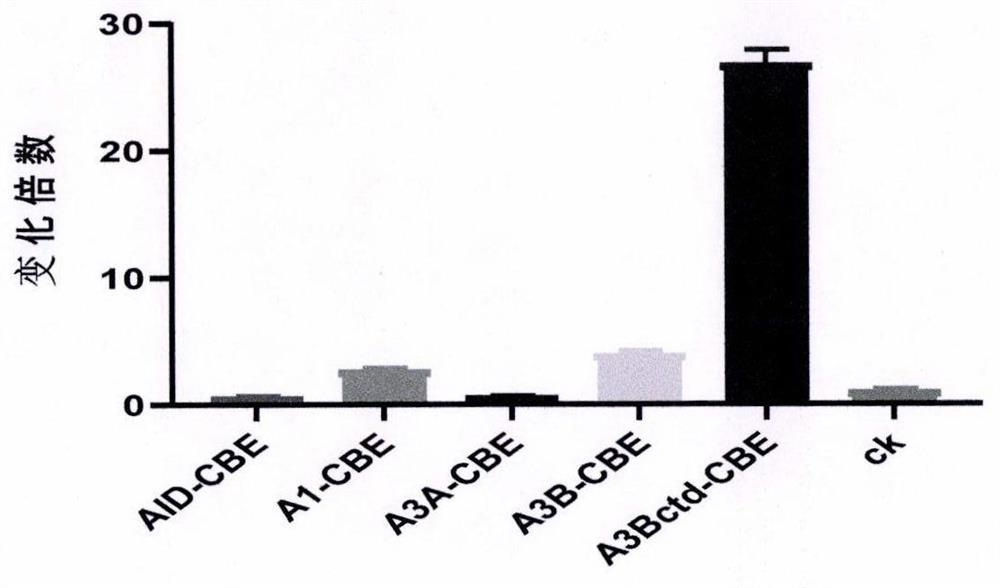
[0044] Example 3. In vitro cytosine editing assays
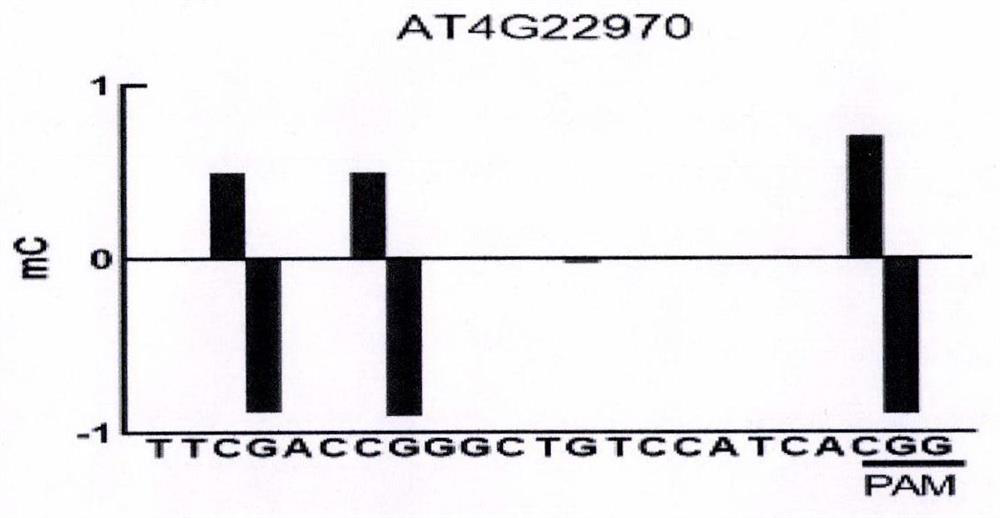
[0045] In this example, different guide RNAs were used to test the cytosine deamination reaction of genomic targets AT4G22970, AT3G13784 and AT1G30950 in vitro.
[0046] Specifically, genomic DNA was extracted from two-week-old Arabidopsis seedlings using DNeasy Plant Maxi Kit (QIAGEN). 500 ng of genomic DNA was reacted with 1 μg of each fusion protein obtained in Example 1 and 1 μg of each guide RNA obtained in Example 2 in 10×CutSmart buffer (NEB) at 37° C. for 3 hours, and the total reaction volume was 20 μl. At the same time, the reaction system without cytosine base editor fusion protein and guide RNA was used as a negative control. After the reaction, use ddH 2 O dilute the reaction system to 40 μl, and add 1 μg of restriction endonuclease to digest at 37°C for at least 8 hours (AT4G22970 target is digested with MspI, the recognition site is CCGG, AT3G13784 target is digested with KpnI, the recognition site The po...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com