Application of magnetic functionalized magnetic nanomaterial and mixed microorganism identification method
A technology of mixing microorganisms and magnetic nanometers, applied in the field of microorganisms, can solve the problems of long-time cultivation, affecting doctors' diagnosis and treatment and patients' recovery time, etc., and achieves the effect of simple operation, problem-solving and auxiliary medical treatment.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1 Enrichment and identification of Escherichia coli and Staphylococcus aureus in different volume ratios in aqueous solution and urine
[0026] Bacterial samples: Escherichia coli (Ec) ATCC 25922, Staphylococcus aureus (Sa) ATCC 25923.
[0027] Specific steps:
[0028] 1. Inoculate the strains frozen at -80°C on Tryptone Soy Agar (TSA) and culture them in a 37°C incubator for 12-14 hours.
[0029] 2. Dissolve the part of the cultured colony in a centrifuge tube filled with sterile water, and the OD of the bacterial solution 600 The value is adjusted to 0.7-0.8. Take a certain volume and put it in another centrifuge tube and centrifuge, then add an equal volume of urine to prepare a specific OD 600 value of monobacterial urine.
[0030] 3. According to the volume ratio of 1:1 and 2:1, take the single bacteria solution and mix them in pairs to obtain the mixed bacteria solution in water and urine with a total volume of 200 μL and 150 μL respectively.
[0031]...
Embodiment 2
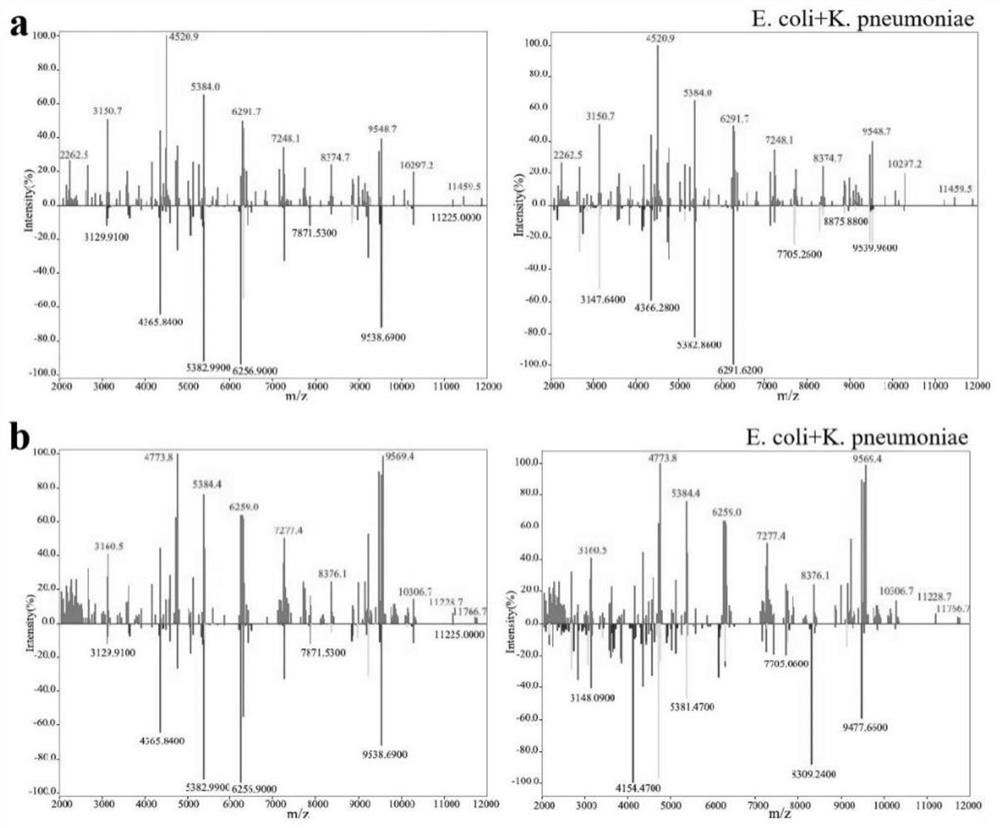
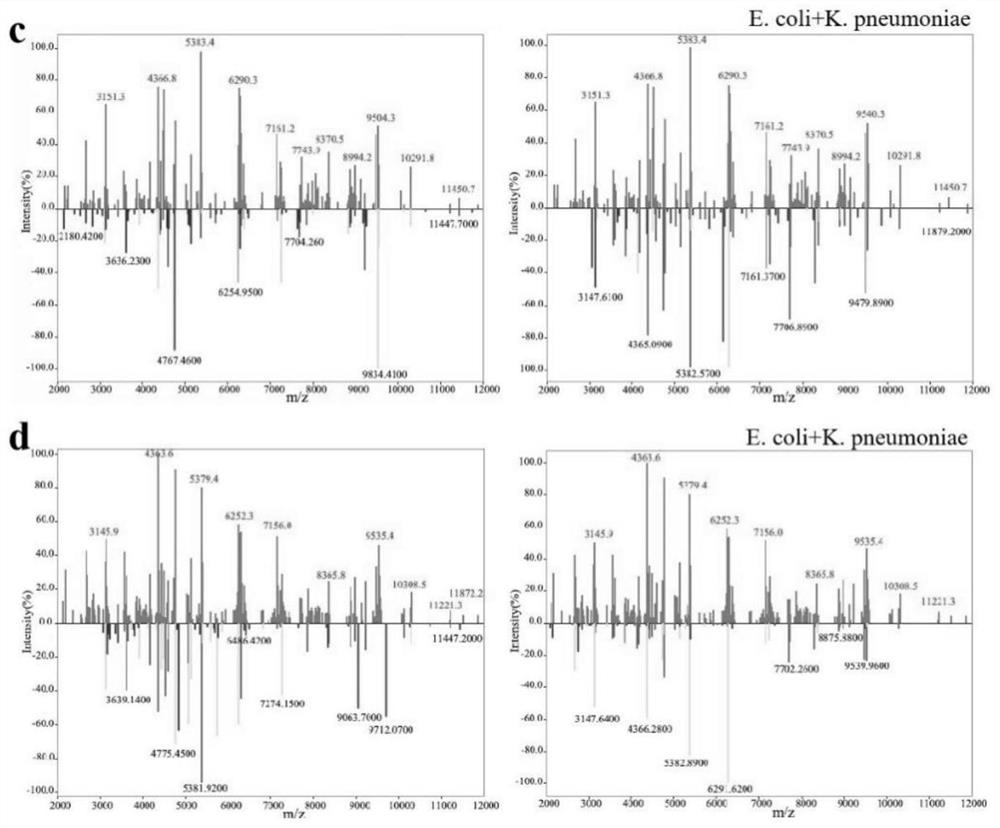
[0039] Example 2 Enrichment and Identification of Escherichia coli and Klebsiella pneumoniae Dimixed Bacteria under Different Volume Ratio in Aqueous Solution and Urine
[0040] Bacterial samples: Escherichia coli (Ec) ATCC 25922, Klebsiella pneumoniae (KP) CICC 21519
[0041] Specific steps:
[0042] 1. Inoculate the strains frozen at -80°C on Tryptone Soy Agar (TSA) and culture them in a 37°C incubator for 12-14 hours.
[0043] 2. Dissolve the part of the cultured colony in a centrifuge tube filled with sterile water, and the OD of the bacterial solution 600 The value is adjusted to 0.7-0.8. Take a certain volume and put it in another centrifuge tube and centrifuge, then add an equal volume of urine to prepare a specific OD 600 value of monobacterial urine.
[0044]3. According to the volume ratio of 1:1 and 2:1, take the single bacteria solution and mix them in pairs to obtain the mixed bacteria solution in water and urine with a total volume of 200 μL and 150 μL respec...
Embodiment 3
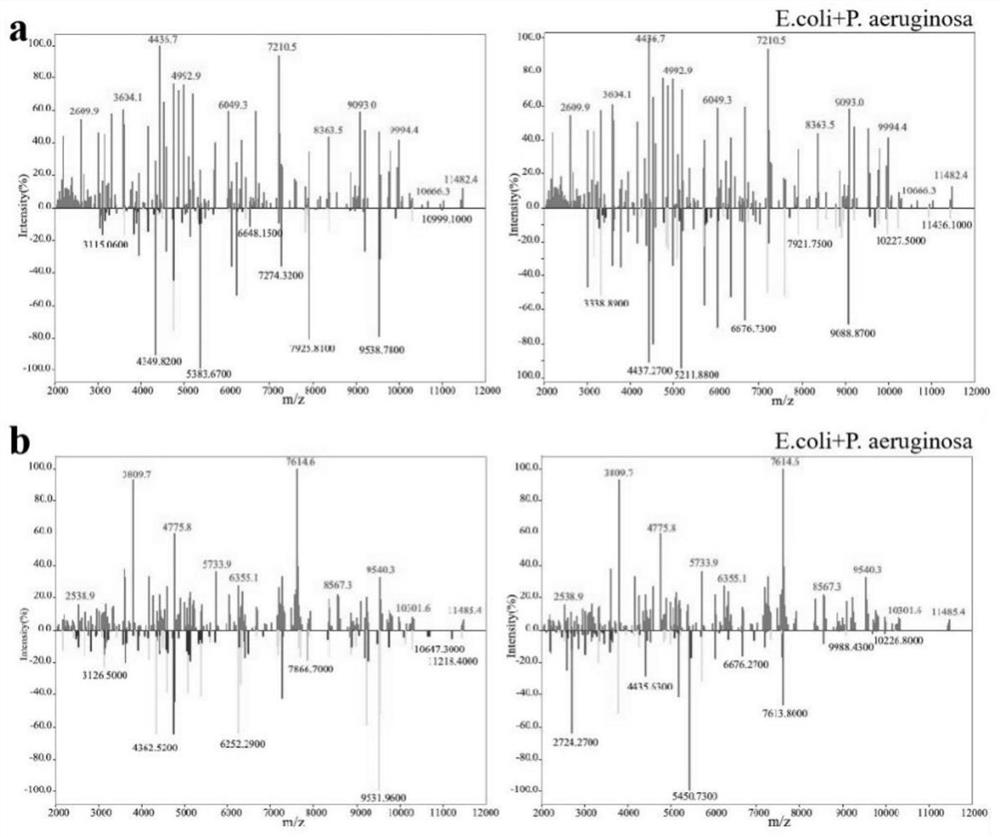
[0053] Example 3 Enrichment and identification of Escherichia coli and Pseudomonas aeruginosa mixed bacteria in different volume ratios in aqueous solution and urine
[0054] Bacterial samples: Escherichia coli (Ec) ATCC 25922, Pseudomonas aeruginosa (PA) CICC 21630 Specific steps:
[0055] 1. Inoculate the strains frozen at -80°C on Tryptone Soy Agar (TSA) and culture them in a 37°C incubator for 12-14 hours.
[0056] 2. Dissolve the part of the cultured colony in a centrifuge tube filled with sterile water, and the OD of the bacterial solution 600 The value is adjusted to 0.7-0.8. Take a certain volume and put it in another centrifuge tube and centrifuge, then add an equal volume of urine to prepare a specific OD 600 value of monobacterial urine.
[0057] 3. According to the volume ratio of 1:1 and 2:1, take the single bacteria solution and mix them in pairs to obtain the mixed bacteria solution in water and urine with a total volume of 200 μL and 150 μL respectively.
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com