Method for rapid quantification of high-throughput sequencing library of MGI platform and kit
A sequencing library, high-throughput technology, used in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., can solve the problems of low specificity, non-specific amplification and inaccurate quantification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
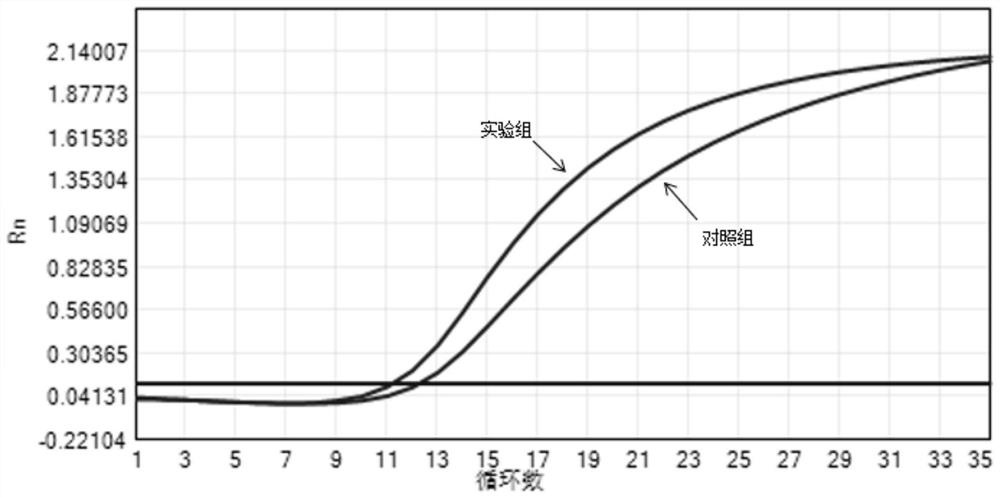
[0056] Embodiment 1 has designed experimental group and control group, wherein experimental group selects SEQ ID NO:1 and SEQ ID NO:2 primer and SEQ ID NO:3 probe for use in experiment group; Control group selects control primer and control probe for use, used contrast The primer and control probe sequences are as follows:
[0057] The control primer includes an upstream primer and a downstream primer, wherein the upstream primer is the sequence shown in SEQ ID NO:9, the downstream primer is the sequence shown in SEQ ID NO:10, and the control probe is the sequence shown in SEQ ID NO:11.
[0058] SEQ ID NO:9CAACTCCTTGGCTCACAGAACG
[0059] SEQ ID NO:10TCCTAAGACCGCTTGGCCTC
[0060] SEQ ID NO:11CATGGCTACGATCCGAC
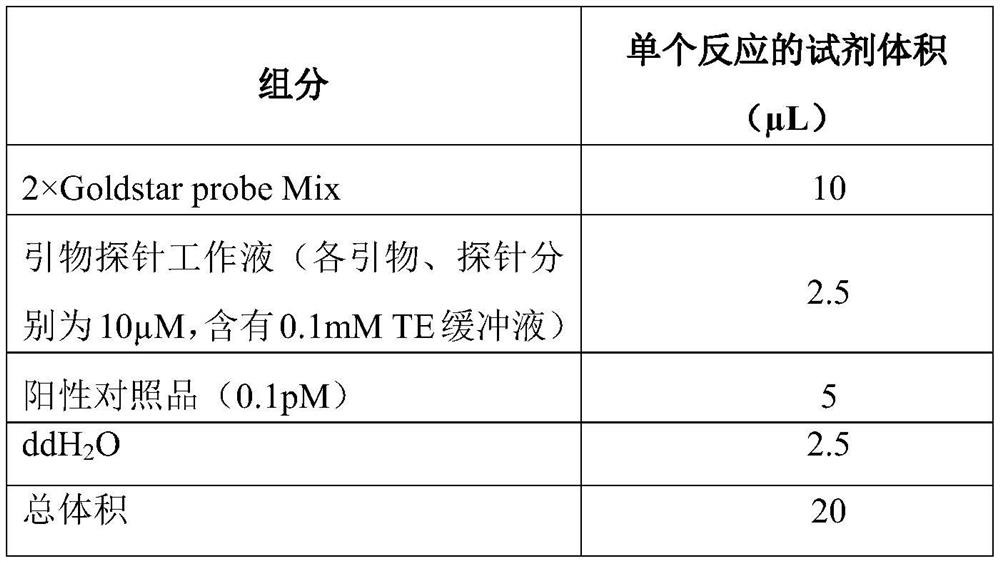
[0061] The above-mentioned primer probes and control primer probes were used to detect the positive standard. The specific PCR amplification system is shown in Table 1, and the amplification conditions are shown in Table 2. Among them, GoldStar Probe Mix is a commer...
Embodiment 2
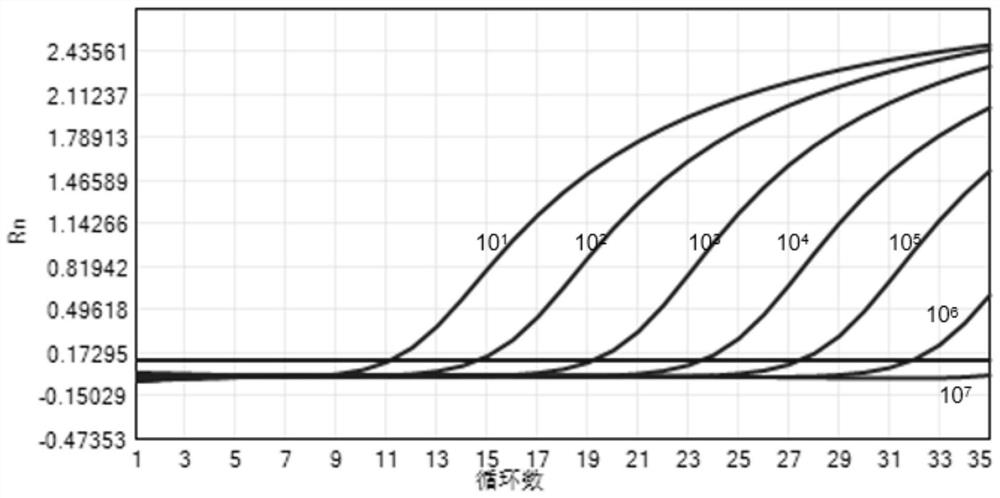
[0068] In order to determine the minimum detection limit of the primers and probes provided by the present invention, firstly, 5 μg of plasmid dry powder was diluted with 100 μL of nuclease-free water to 50 ng / μL as a storage solution, and then the storage solution was serially diluted for 10 1 ~10 8 times, and finally use the primer probe to test the gradient amplification situation to analyze the amplification efficiency and linearity of the primer probe of the present invention.
[0069] Wherein the PCR amplification system is shown in Table 1 of the above-mentioned Example 1, and the amplification conditions are shown in Table 2 of the above-mentioned Example 1. figure 2 Dilute 10 for primer-probe pair 1 ~10 7 The amplification curve results of the positive standard detection times.
[0070] Table 3 is the detection CT value of each concentration positive control substance for the provided primer probe
[0071] group Detection of CT value Dilution 10 ...
Embodiment 3
[0074] In the present invention, the Arabidopsis thaliana PHYA gene fragment is selected as an internal standard, and multiple fluorescent PCR technology is used to quantify the library and at the same time control the quality of the amplification reaction process. In this embodiment, the internal standard exists in the library diluent, and the working concentration is 0.1 pM. On the one hand, it can be used as the diluent of the library, and at the same time, it can also realize the function of the internal standard.
[0075] Example 1 selects 10 libraries qualified for quality inspection as amplification templates. The sample types include alveolar lavage fluid, sputum, blood, cerebrospinal fluid, tissues, etc., by means of fluorescent PCR (reaction system and conditions refer to the table in the embodiment) 1 and Table 2) for detection. The primer probe working solution also contains 10 μM internal standard upstream primer SEQ ID NO:4, 10 μM internal standard downstream pri...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com