Method for accurately positioning chromosome deletion/repetition breakpoint
A chromosomal deletion and precise positioning technology, applied in biochemical equipment and methods, recombinant DNA technology, and microbial assay/inspection, etc., can solve the problems of complex operation, high cost, long detection cycle, etc., and achieve high efficiency and low cost. , the effect of design science
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] This embodiment discloses a case of DMD gene No. 53-55 exon 53-55 exon deletion breakpoint roughly regional positioning, specifically:
[0041] 1. Use nucleic acid extraction or purification reagents from Nahai Hi-Tech Biotechnology Co., Ltd. to extract the patient's genomic DNA.
[0042] 2. Using restriction endonuclease (or sonication) to fragment the genomic DNA sample into fragments with an average length of about 150-250 bp. The fragment was then subjected to end repair and an "A" was added.
[0043] Fragmentation, end repair, and the conditions for adding A:
[0044] 37℃ 40min
[0045] 65℃ 30min
[0046] 4℃ hold
[0047] 3. Then connect adapters with tag sequences to both ends,
[0048] The connection condition is: 20°C 15min.
[0049] 4. Then, perform PCR to enrich the library, and the PCR conditions are:
[0050] Pre-denaturation at 95°C for 3 minutes
[0051] (denaturation at 98°C for 20s, annealing at 60°C for 15s, extension at 72°C for 30s) 7 cycles a...
Embodiment 2
[0063] This embodiment discloses an example of the accurate location of the breakpoint of exon 53-55 deletion of DMD gene, specifically:
[0064] 1. Through data analysis, the sample in Example 1 has exon 53-55 deletion, the upstream breakpoint range is chrX: 31856593-31863252, the downstream breakpoint range is chrX: 31954056-31955026, in the above suspected breakpoint area Upstream and downstream Design 13 upstream primers and one universal downstream primer; specifically:
[0065] DMD-1F GGGCAGCAAAGGGAAGATAAATA, as shown in SEQ ID NO:1;
[0066] DMD-2F TGTGATAAAAGTGCAAAAGAGTTCGC, as shown in SEQ ID NO:2;
[0067] DMD-3F GGCCCATCTGAATAGTCCATAAAATC, as shown in SEQ ID NO:3;
[0068] DMD-4F GTCCTTATAGTTTCTCATTTCTCTGC, as shown in SEQ ID NO:4;
[0069] DMD-5F ATATTCCTCCACAAGACATTCCAGTG, as shown in SEQ ID NO:5;
[0070] DMD-6F GATTTGTAATGCCCTATGGACCAGA, as shown in SEQ ID NO:6;
[0071] DMD-7F TTGCAGTCTTTTGTGTCAGTGTAGA, as shown in SEQ ID NO:7;
[0072] DMD-8F ACGGTAGGAGT...
Embodiment 3
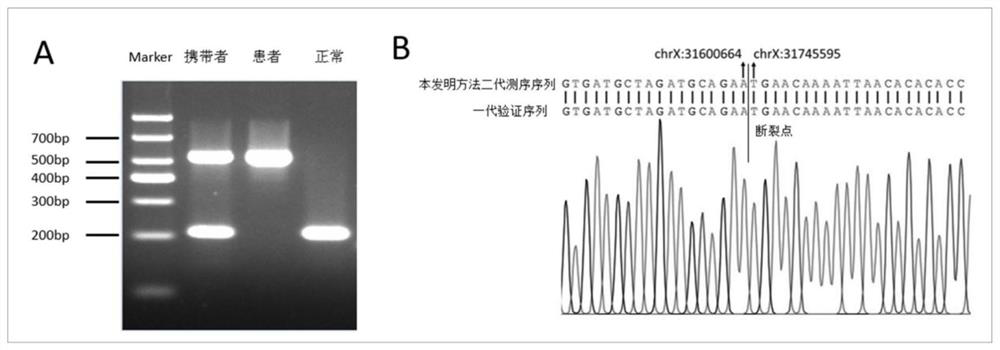
[0104] This embodiment discloses the verification of the DMD gene exon 53-55 deletion breakpoint of embodiment 2, specifically:
[0105] According to the confirmed breakpoint position, primers were designed on the flanks, and primers were designed inside the deletion region to amplify the normal non-deleted sequence. The length of the product inside the breakpoint was 209bp, and the length of the product outside the breakpoint was 533bp. 100ng) genomic DNA was subjected to PCR and then run agarose gel electrophoresis,
[0106] PCR conditions:
[0107] Pre-denaturation at 94°C for 5 minutes
[0108] (denaturation at 94°C for 30s, annealing at 60°C for 30s, extension at 72°C for 3min) 38 cycles and final extension at 72°C for 10min
[0109] Store at 4°C hold
[0110] Agarose gel electrophoresis conditions:
[0111] 3% agarose, electrophoresis at 50V for 120min.
[0112] Carriers should be two bands of different lengths, patients should be one longer band, and negative contr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com