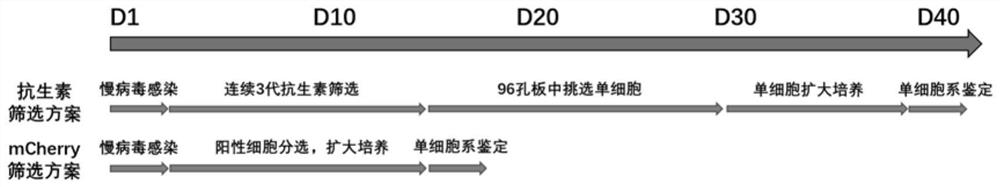
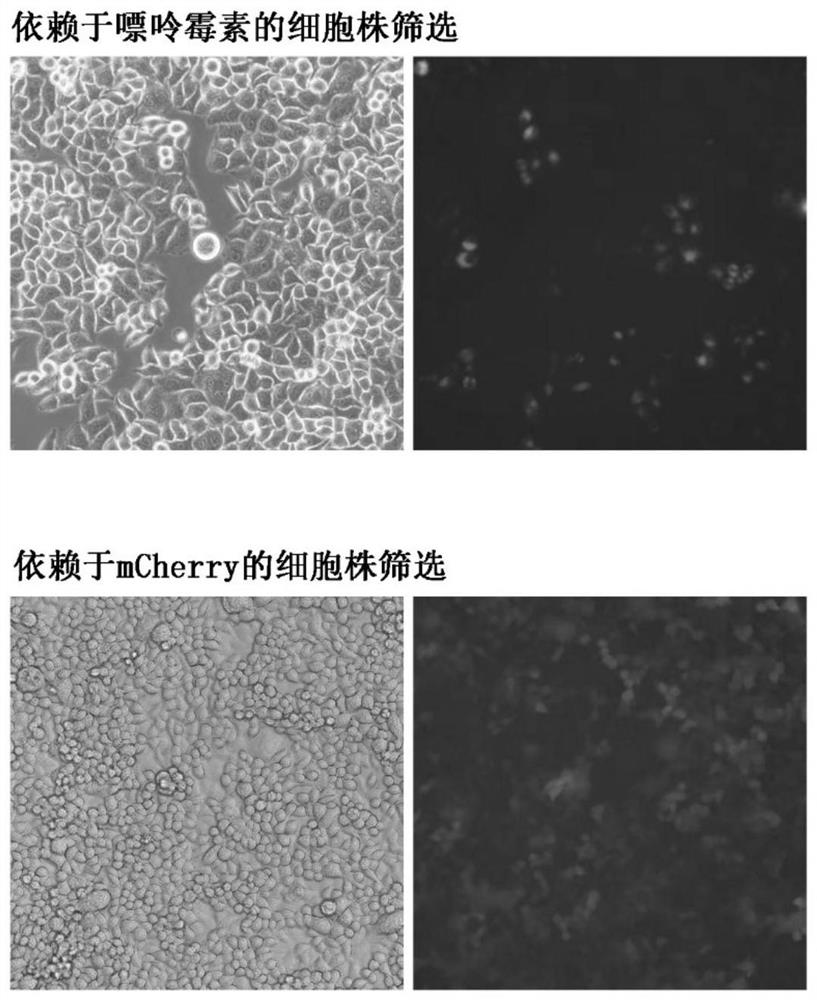
Detection method of content of neutralizing antibody of adeno-associated virus and construction method of cell line
A construction method and detection method technology, applied in the field of medical detection, can solve the problems of inability to meet the real-time monitoring of stable cell lines, long construction period, false positives of cell lines, etc., so as to avoid the problem of drug-resistant bacteria and shorten the construction time.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] This embodiment provides a method for constructing a cell line sensitive to adeno-associated virus infection, and the specific construction steps are as follows:
[0061] Introduce the lentiviral vector overexpressing the miniAAVR-mCherry gene into Hela cells; obtain Hela cells overexpressing miniAAVR-mCherry, that is, Hela cells sensitive to adeno-associated virus infection;
[0062] Adeno-associated virus-infected sensitive Hela cells were cultured to obtain a large number of adeno-associated virus-infected sensitive Hela cells.
[0063] Specifically, the construction process of the lentiviral vector of miniAAVR-mCherry gene is as follows (linear sequence refers to image 3 shown):
[0064] (1) Original plasmid sequence:
[0065] AAVR cDNA: Extract RNA from hela cells, reverse transcribe cDNA, use this as a template, and PCR produce AAVRcDNA.
[0066] Vector sequence: the vector is pLVX-Puro vector, purchased from transvector;
[0067] mCherry sequence: CDS region...
Embodiment 2
[0115] This embodiment provides a method for detection and content determination of adeno-associated virus neutralizing antibodies, and the specific operation steps are as follows:
[0116] The adeno-associated virus infection-sensitive Hela cells constructed in Example 1 were cultured in a 24-well plate, and the number of cells was 1×10 5 Cells / well density were plated;
[0117] Dilute the sample to be tested in a 10-fold concentration gradient, the highest dilution is 1×10 9 times, to obtain 10 different concentrations of samples to be tested;
[0118] And set the maximum value control group (do not contain AAV antibody, add neonatal umbilical cord blood serum to replace the antibody sample) and minimum value control group (do not add AAV during detection, use DMEM to replace AAV);
[0119] Take 10 samples of different concentrations to be tested, the maximum value control group and the minimum value control group, a total of 12 groups, take 10 μL respectively, mix and inc...
Embodiment 3
[0124] This embodiment provides a method for detection and content determination of adeno-associated virus neutralizing antibodies, and the specific operation steps are as follows:
[0125] (1) The adeno-associated virus infection-sensitive Hela cells constructed in Example 1 were cultured in a 24-well plate, and the number of cells was 1×10 5 Cells / well density were plated;
[0126] (2) Dilute the sample to be tested in a 10-fold concentration gradient, the highest dilution is 1×10 9 times, to obtain 10 different concentrations of samples to be tested;
[0127] (3) And set the maximum value control group (without AAV antibody, adding neonatal umbilical cord blood serum to replace the antibody sample) and the minimum value control group (do not add AAV during detection, use DMEM instead of AAV);
[0128] (4) Take 10 different concentrations of samples to be tested, the maximum value control group and the minimum value control group, a total of 12 groups, take 30 μL respectiv...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com