Somatic mutation hypersensitivity detection method based on nucleic acid mass spectrum platform
A nucleic acid mass spectrometry and detection method technology, which is applied in the field of ultrasensitive detection of somatic mutation based on a nucleic acid mass spectrometry platform, can solve the problems of easy pollution, long time and high cost, and achieves wide application, short detection time and low cost. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
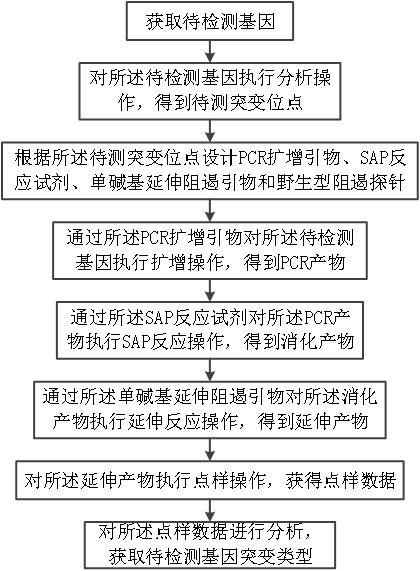
[0073] Embodiments of the present invention provide a method based on nucleic acid mass spectrometry platform cell mutation hypermissone detection method, please refer to Figure 1 to 22 , Includes the following steps:
[0074] It is to be noted that the selected test site is L858R and T790M, and the sequence information of the design is selected by the EGFR. Image 6 Indicated.
[0075] The gene EGFR to be detected was obtained, and the sequence analysis operation was performed, i.e., the region DNA sequence of the EGFR gene was analyzed, and the mutagenection site L858R and T790M were obtained to be tested.
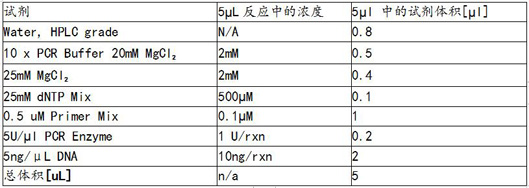
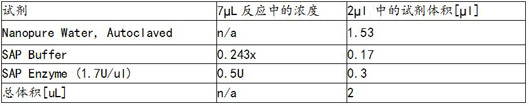
[0076] The reaction system of the PCR amplification probe according to the EGFR gene is designed, SAP reaction reagent, single base extension repressor and wild type repressor, and PCR amplification probes. figure 2 As shown, the reaction system of the SAP reactant is as image 3 The reaction system of the single base extension of the plasma Figure 4 As shown, the design of wi...
Embodiment 2
[0097] Embodiments of the present invention provide a method based on nucleic acid mass spectrometry platform cell mutation hypermissone detection method, please refer to Figure 1 to 5 and Figure 23 to Figure 29 , Includes the following steps:
[0098] It is to be noted that the selected test site is G12D and G13D, and the sequence information of the design is selected as the KRAS. Figure 23 Indicated.
[0099] The gene KRAS to be detected is obtained, and the sequence analysis operation is performed, that is, the region DNA sequence of the KRAS gene is analyzed, and the mutagenection point G12D and G13D to be tested are obtained.
[0100] According to the KRAS gene, the PCR amplification primer, SAP reagent, mono-base extension repressor and wild-type repressor probe, the reaction system of PCR amplification primers, based on the KRAS gene.figure 2 As shown, the reaction system of the SAP reactant is as image 3 The reaction system of the single base extension of the plasma Figure...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com