Microorganism contrast result correction method and system based on metagenome sequencing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
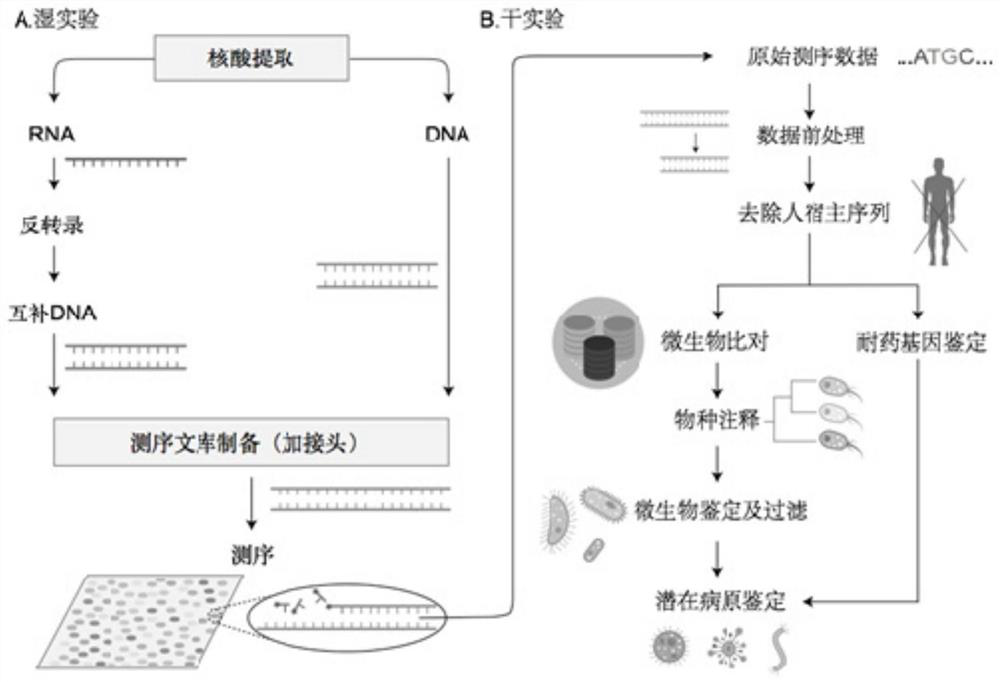
[0094] Embodiment 1 System and method for detecting microorganisms in a sample
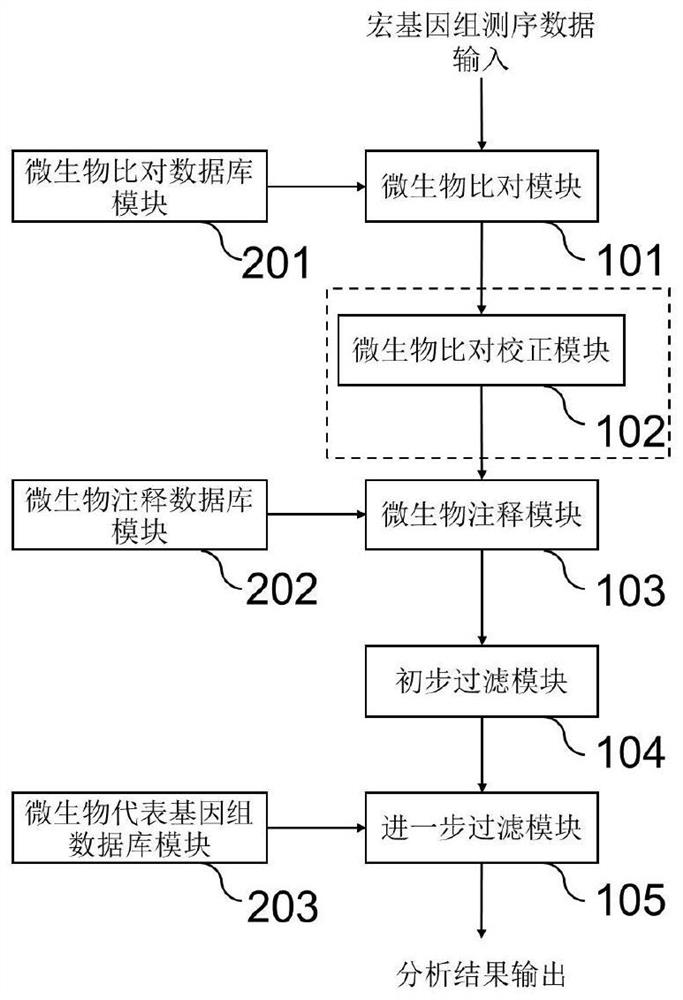
[0095] This embodiment provides a system for detecting microorganisms in a sample, such as figure 1 shown, including:
[0096] Microbial analysis module set, including: microbial comparison module 101, microbial annotation module 103, preliminary filtering module 104 and further filtering module 105; and
[0097] The microbial database module group includes: a microbial comparison database 201, a microbial annotation database 202 and a microbial representative genome database 203,
[0098] in,
[0099] The microbial comparison module 101 is connected to the microbial comparison database 201, and is used to compare and analyze the metagenomic sequencing data of the sample based on the K-mer algorithm using the microbial comparison database to obtain the microbial comparison result;
[0100] The microorganism annotation module 103 is connected with the microorganism comparison module 101 and the ...
Embodiment 2
[0110] Embodiment 2 System and method for detecting microorganisms in a sample
[0111] This embodiment improves the system for detecting microorganisms in samples in Example 1. The point of improvement is that the microbial analysis module set further includes a microbial comparison correction module set 102 (such as figure 1 inside the dotted line box), located between the microorganism comparison module 101 and the microorganism annotation module, for correcting the microorganism comparison result obtained in S21 based on the following steps.
[0112] Search by family as a unit, if the sum of the sequences annotated to each genus accounts for less than 50% of the total sequence of the family, and the relative abundance of the family is greater than 15%, then:
[0113] a) Extract and assemble the reads annotated to the family into a contig;
[0114] b) compare reads to contig, and record the corresponding relationship between read and contig;
[0115] c) compare the contig...
Embodiment 3
[0123] Embodiment 3 Detection system and method for microorganisms and drug-resistant genes in samples
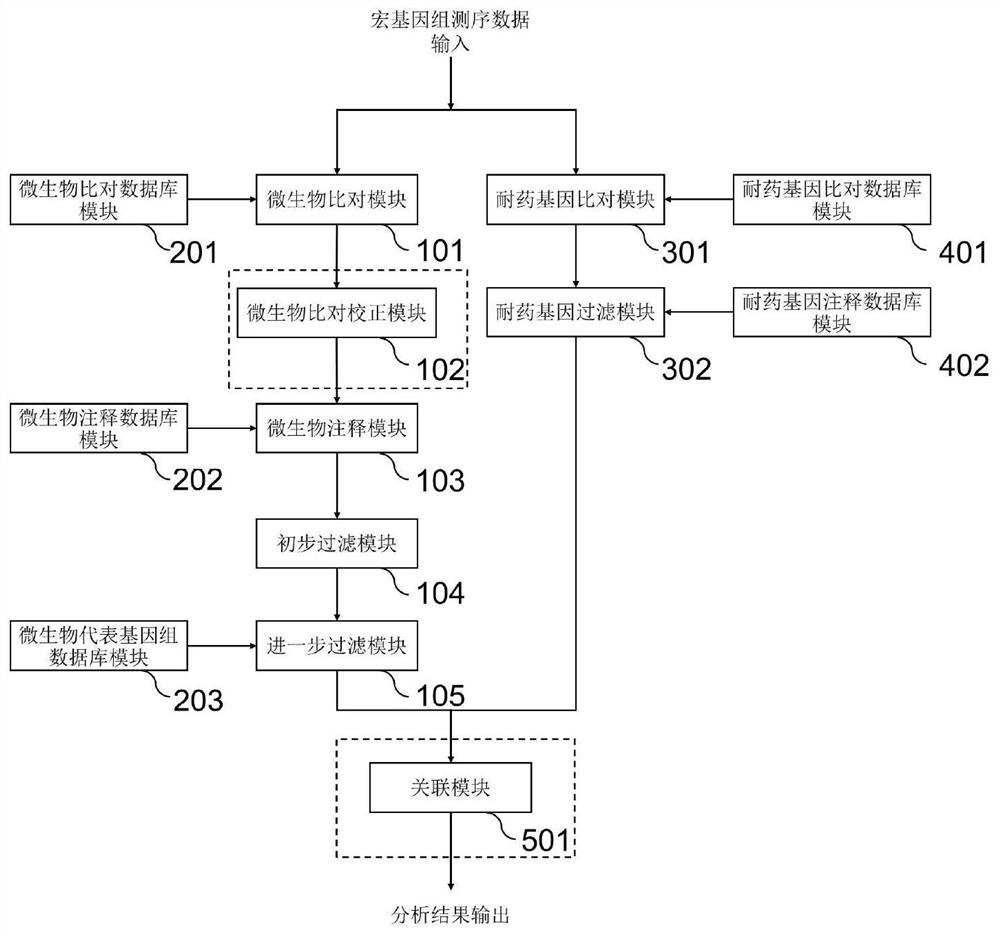
[0124] This embodiment improves the system for detecting microorganisms in the sample of embodiment 1 or embodiment 2, such as figure 2 As shown, the point of improvement is to further include:
[0125] The drug-resistant gene analysis module set includes: a drug-resistant gene comparison module 301 and a drug-resistant gene filter module 302;
[0126] Drug-resistant gene database module group: including: drug-resistant gene comparison database module 401 and drug-resistant gene annotation database 402,
[0127] in,
[0128] The drug-resistant gene comparison module 301 is connected to the drug-resistant gene comparison database module 401, and is used to compare and analyze the metagenomic sequencing data of samples using the drug-resistant gene database to obtain predicted drug-resistant gene information;
[0129] The drug-resistant gene filtering module 302 is respec...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com