SNP molecular marker for column number of secondary milk ducts of rubber trees and application of SNP molecular marker
A rubber tree and marker technology, which is applied in the determination/inspection of microorganisms, recombinant DNA technology, biochemical equipment and methods, etc., can solve problems such as not involving cultivated varieties, improve breeding efficiency, shorten selection cycle, and reduce breeding workload. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1: Acquisition of SNP markers related to the number of secondary milk duct rows
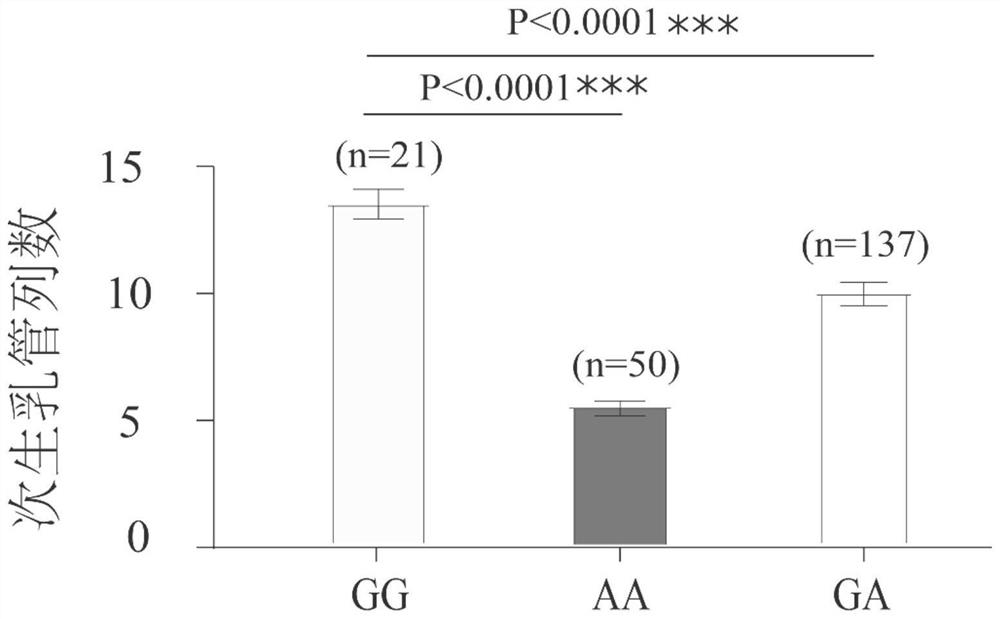
[0025] The number of secondary milk ducts in cultivars of rubber trees was significantly more than that in wild species. The materials used in this study are 208 germplasm resources of rubber trees preserved in the National Rubber Tree Germplasm Resource Garden (Danzhou, Hainan), including 93 cultivated species and 115 wild species. Fresh leaves were obtained, genomic DNA was extracted by CTAB method, and whole genome resequencing was performed. GATK software was used to obtain the variation data of all materials, and the SNP differences between wild species and cultivated species were analyzed, and a SNP site related to the number of secondary laticiferous ducts of rubber trees was identified.
[0026] The site is located at the 238bp site from the 5' end of the sequence shown in SEQ ID NO: 1, and the site is represented by "g" in the sequence of SEQ ID NO: 1, and the base of the...
Embodiment 2
[0027] Example 2: Evaluation of GG genotype and AA genotype on missed selection rate and misselection rate of germplasm with large number of duct rows
[0028] 1. Experimental materials
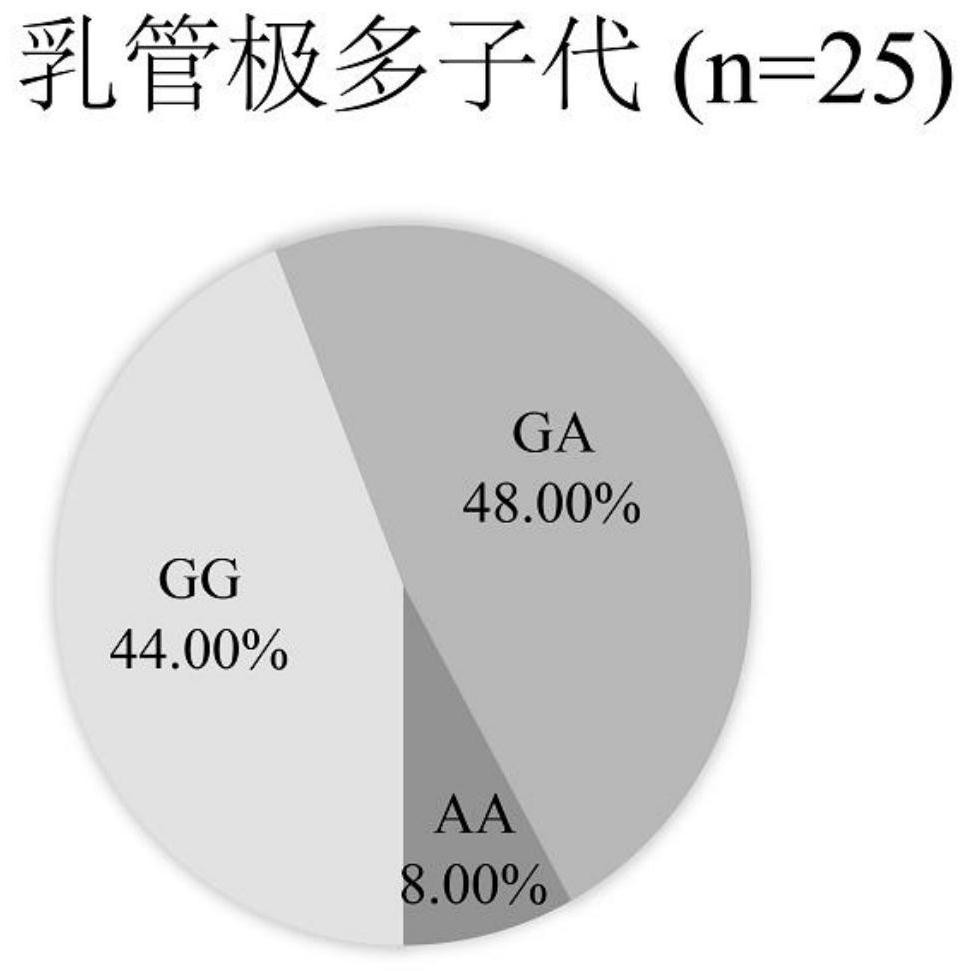
[0029] The laticifer differentiation ability of 286 hybrid progenies with unknown pedigree that were planted in the ninth team of the Institute of Rubber, Chinese Academy of Tropical Agricultural Sciences was evaluated, and 25 progenies with a very large number of secondary laticifer columns (more than 12 columns) were selected to obtain fresh leaves Genomic DNA was extracted according to the instructions of the Plant Genomic DNA Extraction Kit (Tiangen Biochemical Technology Co., Ltd.), and the extracted DNA was tested by 1.5% agarose gel electrophoresis and NanoDrop 2000 for later use.
[0030] 2. PCR amplification
[0031] PCR amplification was carried out using the extracted genomic DNA as a template, the sequence of the forward primer used was SEQ ID NO.2, and the sequence of the revers...
Embodiment 3
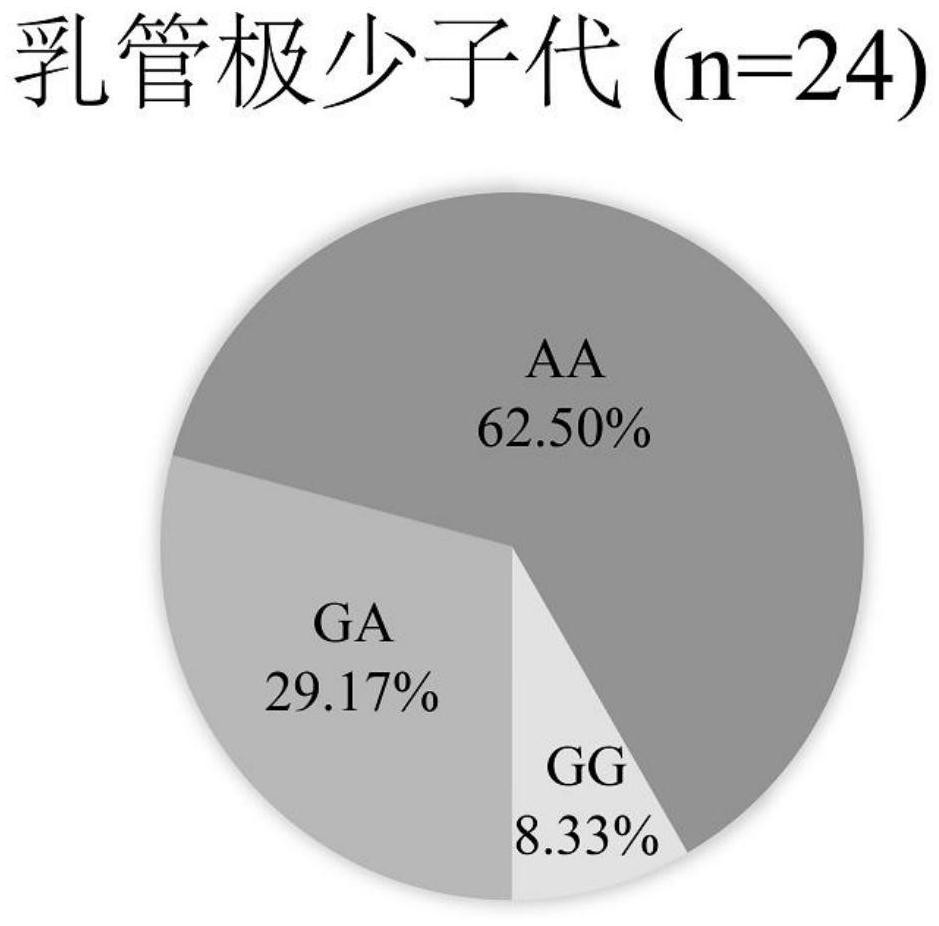
[0036] Example 3: Evaluation of AA genotype and GG genotype on missed selection rate and misselection rate of germplasm with few milk duct rows
[0037] 1. Experimental materials
[0038] The 286 progenies of 7 hybrid combination populations were evaluated for laticiferial differentiation ability (same as Example 2), and 24 progenies with a very small number of secondary laticifer columns (less than 3 columns) were selected to obtain fresh leaves. According to the plant genome DNA Genomic DNA was extracted according to the instructions of the extraction kit (Tiangen Biochemical Technology Co., Ltd).
[0039] 2. PCR amplification
[0040] PCR amplification was carried out using the extracted genomic DNA as a template, the sequence of the forward primer used was SEQ ID NO.2, and the sequence of the reverse primer was SEQ ID NO.3. The PCR amplification system is 20ul: 10 μL of 2x PCR buffer, 1 μL of forward and reverse primers (10 μmol L-1), 1 μL of DNA template, ddH 2 O is 7 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com