SNP molecular marker combination for identification of genetic relationship of pigs, application and method
A technology of kinship and molecular markers, applied in the field of molecular genetics, to achieve accurate and reliable typing results, complete and accurate pedigree, and guaranteed accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
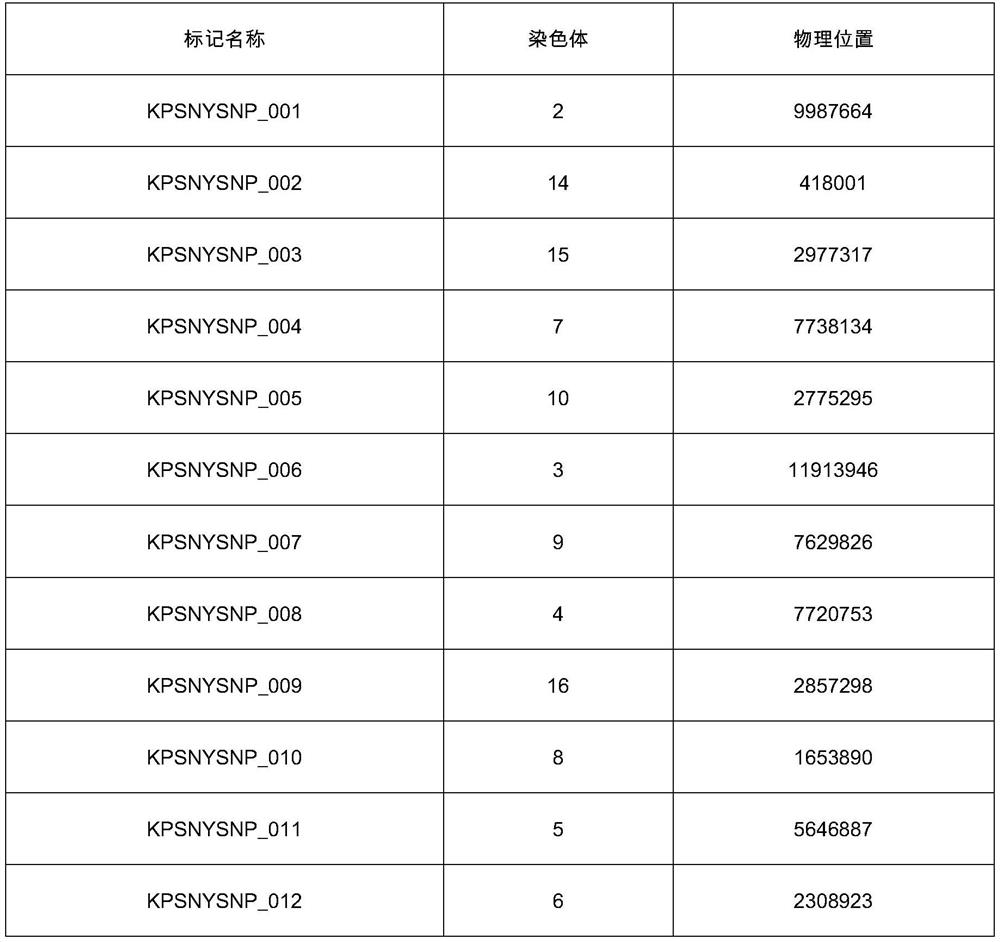
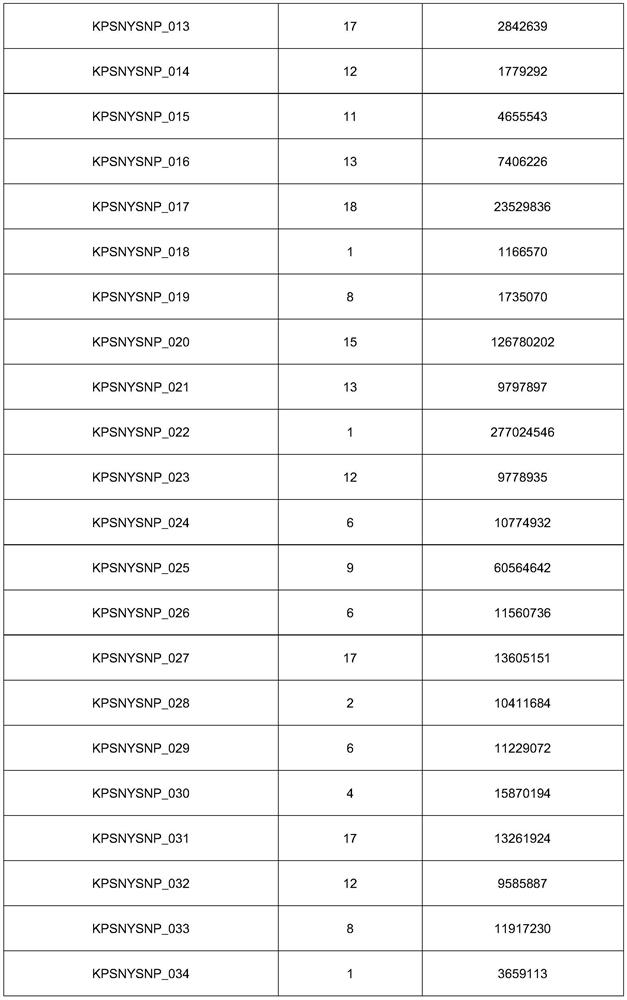
[0263] Embodiment 1: the selection of SNP marker combination;
[0264] The present invention has gone through 3 processes for the SNP marker selection of the paternity identification system for Du, Chang, and large commercial breed pig herds, specifically as follows:
[0265] (1) Group division
[0266] The Du, Long and large commercial pig populations were divided into marker screening population and marker verification population, the screening population was used for screening and verification, and the marker verification population was only used for marker verification.
[0267] (2) Marker screening
[0268] For the genotype data of CMIC-1 in different populations, firstly, according to callrate (>0.95), Harwin equilibrium (p>10 -6 ) to filter out the SNP markers that do not meet the requirements, and then screen out the SNP markers with MAF>0.3. Finally find out the intersection of SNP markers that meet the requirements of all groups. Finally, LD filtering was perform...
Embodiment 2
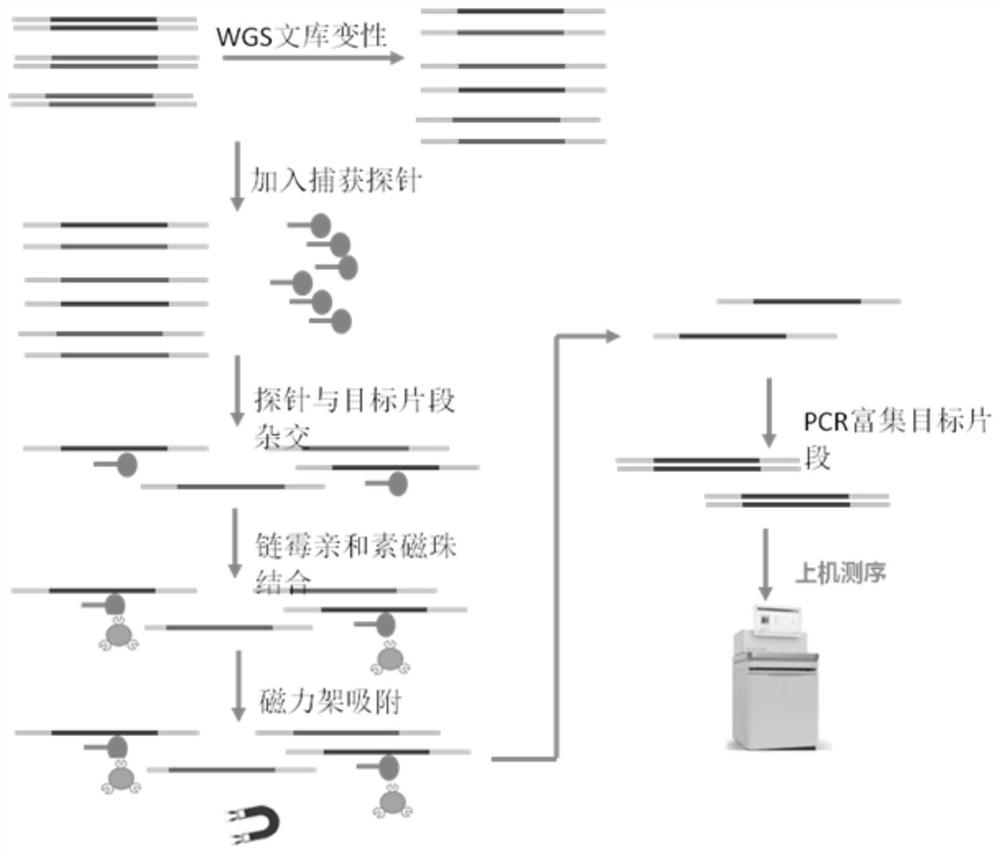
[0280] Embodiment 2: Sampling and targeted capture sequencing technology SNP polymorphism analysis and paternity identification and inference are carried out in pig herds; specifically, the following steps are included:
[0281] Step S1, preparing a porcine 200SNP-labeled liquid chip based on targeted capture sequencing;
[0282] In this step, the porcine 200SNP-labeled liquid chip is composed of independently packaged probes and hybrid capture reagents. The pig 200SNP labeled probe is a single-stranded RNA probe, which is a nucleotide sequence designed and synthesized according to the screened SNP site. The design principle of the probe is that the length of the probe is 120bp, the GC content of the probe is between 25% and 75%, and the number of homology comparisons is less than or equal to 10. According to the SNP sites obtained by screening, two oligonucleotide sequences covering the SNP sites were designed, and the 5' short nucleotide sequences were modified with biotin ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com