Genetic transformation method of rehmannia glutinosa libosch
A genetic transformation method, Tianmu Dihuang technology, applied in biochemical equipment and methods, botanical equipment and methods, and the use of vectors to introduce foreign genetic materials, etc., can solve the problems of low genetic transformation efficiency and large genetic differences, and achieve healing Reduced browning rate, increased regeneration and differentiation rate, and convenient phenotypic identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
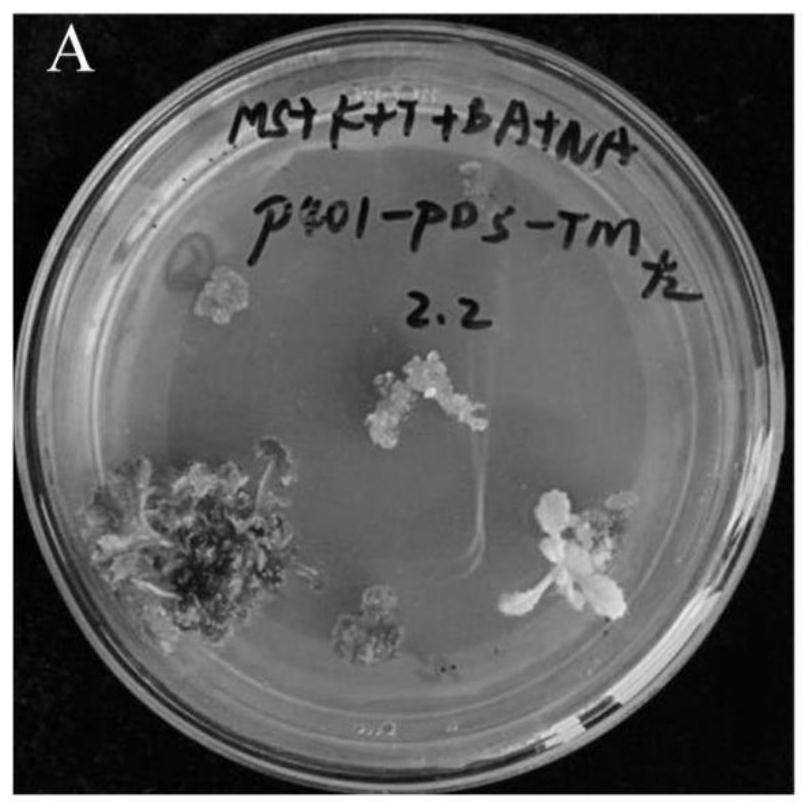
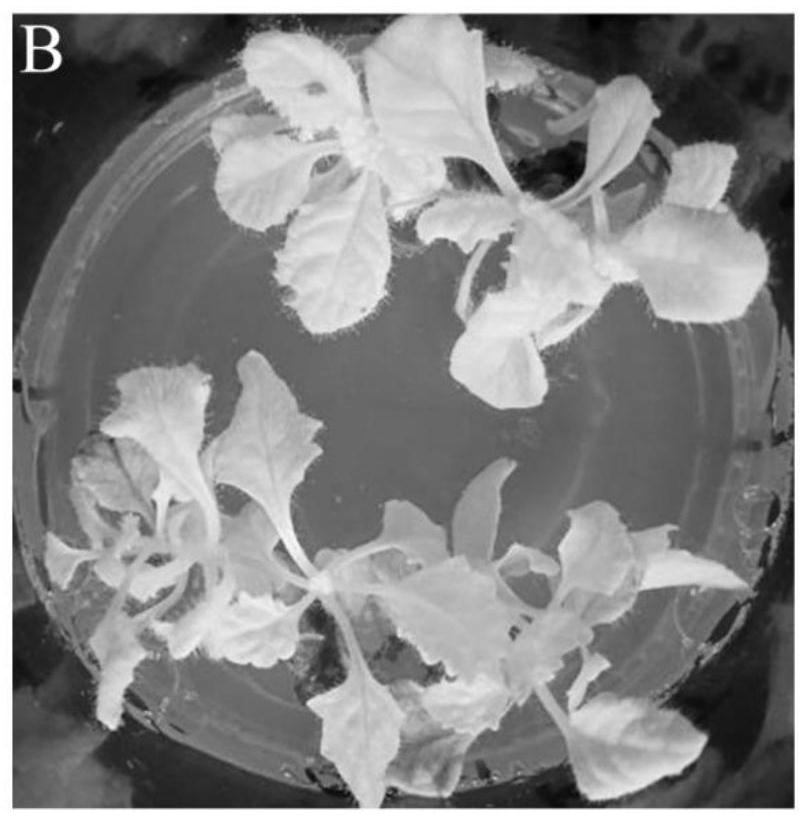
Examples
Embodiment 1
[0043]A method for genetic transformation of Rhizoma Rehmanniae, comprising:
[0044] Cloning of PDS Gene of Tianmu Dihuang
[0045] The young leaves of Tianmu Dihuang were selected to extract total RNA, and the extraction method was carried out with reference to the instructions of the RNA extraction kit (TaKaRa, Dalian);
[0046] Reverse transcription was performed using cDNA first-strand synthesis kit, and the method was carried out with reference to the reverse transcription kit type 6210A (TaKaRa, Dalian) kit instructions;
[0047] The transcript sequence encoding phytoene dehydrogenase (PDS) was screened in the Tianmu Dihuang transcriptome, and it was predicted to have a complete open reading frame (ORF), which was named RcPDS, and its nucleotide sequence is shown in SEQ ID NO. 1 mentioned;
[0048] According to the sequence of RcPDS, a pair of specific primers RcPDS_F and RcPDS_R were designed, and the cDNA synthesized by reverse transcription was used as a template, ...
Embodiment 2
[0072] A method for genetic transformation of Tianmu Rehmannia, comprising the following steps:
[0073] (1), Tianmu Dihuang PDS Gene Cloning
[0074] According to the transcriptome annotation results of Rehmannia glutinosa, a transcript with the highest homology to RgPDS1 gene of Rehmannia glutinosa was screened, specific PCR primers were designed, and the cDNA of Rehmannia glutinosa was used as a template to amplify the PDS gene of Rehmannia glutinosa with high-fidelity DNA polymerase The full-length coding sequence, the nucleotide sequence of its coding region is shown in SEQ ID NO.1;
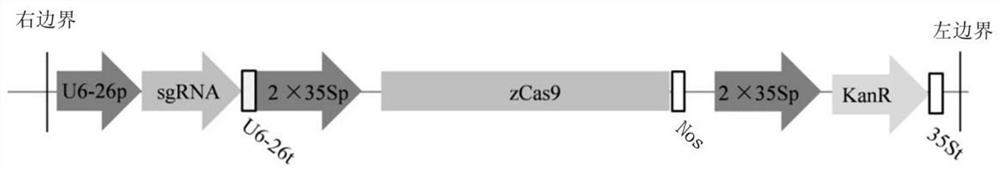
[0075] (2), CRISPR / Cas9 gene editing vector construction and loaded into Agrobacterium tumefaciens
[0076] For the PDS gene obtained in the cloning step (1), select a target sequence that can be efficiently edited by the Cas9 gene at the end of the exon close to the ATG, and insert the sgRNA target sequence into the CRISPR / Cas9 gene editing vector by using a mixed restriction enzyme ligati...
Embodiment 3
[0096] A method for genetic transformation of Tianmu Rehmannia, comprising the following steps:
[0097] (1) Cloning of Tianmu Dihuang PDS Gene
[0098] According to the transcriptome annotation results of Rehmannia glutinosa, a transcript with the highest homology to RgPDS1 gene of Rehmannia glutinosa was screened, specific PCR primers were designed, and the cDNA of Rehmannia glutinosa was used as a template to amplify the PDS gene of Rehmannia glutinosa with high-fidelity DNA polymerase The full-length coding sequence, the nucleotide sequence of its coding region is shown in SEQ ID NO.1;
[0099] (2), CRISPR / Cas9 gene editing vector construction and loaded into Agrobacterium tumefaciens
[0100] For the PDS gene obtained in the cloning step (1), select a target sequence that can be efficiently edited by the Cas9 gene at the end of the exon close to the ATG, and insert the sgRNA target sequence into the CRISPR / Cas9 gene editing vector by using a mixed restriction enzyme liga...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com