EST-SSR (expressed sequence tag-simple sequence repeat) molecular marker, kit and identification method for identifying mesona chinensis variety
A technology of molecular markers and kits, applied in biochemical equipment and methods, microbe determination/testing, DNA/RNA fragments, etc., to achieve high reliability, high resolution, and better resolution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] The inventor team designed and synthesized 192 pairs of SSR primers based on the transcriptome data of jelly grass, and screened the identification effect of jelly jelly material. The statistics of some primer sets and identification results are as follows:
[0046]
[0047]
[0048]
[0049]
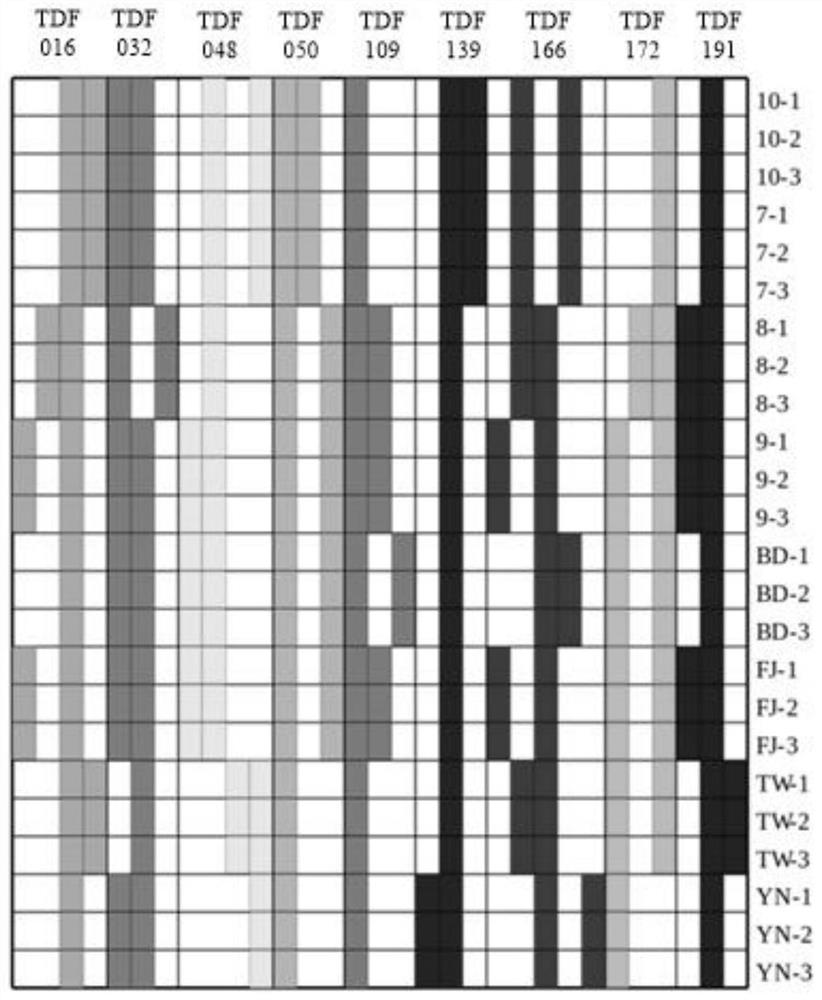
[0050] Through repeated experiments, it was found that the identification effect of most of the SSR primers designed and synthesized based on transcriptome data was not good. Only the 9 sets of primers shown in the following table are used for better identification of the sites of Xiancao varieties, and the information of the relevant sites and their primer sets is shown in the table below:
[0051]
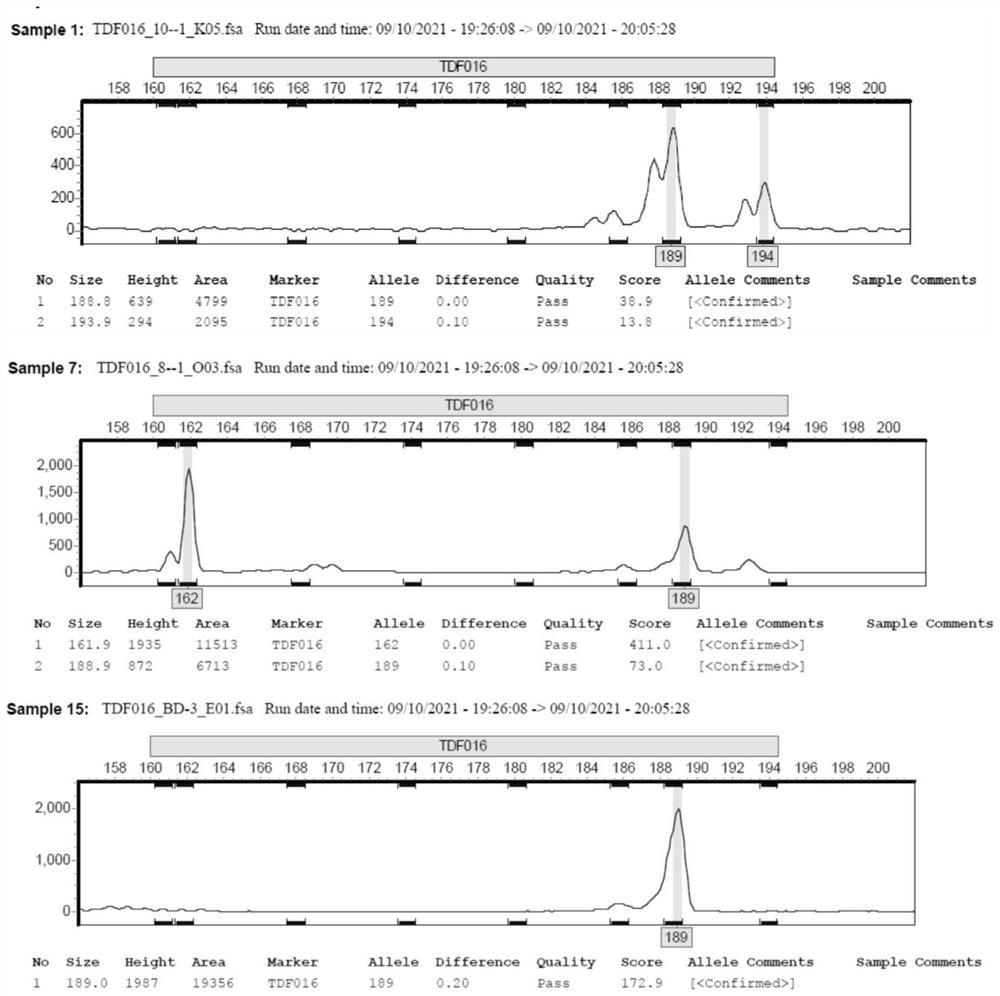
[0052]Further, the genetic diversity analysis for loci: Na, Ne, I, Ho, He, F, Fst and other parameters were calculated using GenAIex software, and the PIC value of each locus was calculated by Cervus software. The following are the specific analysis results and their ...
Embodiment 2
[0057] The present embodiment provides the identification method of celestial grass kind, comprises the steps:
[0058] S1, provide samples to be tested: BD-1, BD-2, BD-3, FJ-1, FJ-2, FJ-3, YN-1, YN-2, YN-3, TW-1, TW-2 , TW-3, 7-1, 7-2, 7-3, 8-1, 8-2, 8-3, 9-1, 9-2, 9-3, 10-1, 10-2, 10 -3. Please refer to the table below for information on the source of samples of different varieties:
[0059]
[0060]
[0061] S2, extraction of genomic DNA:
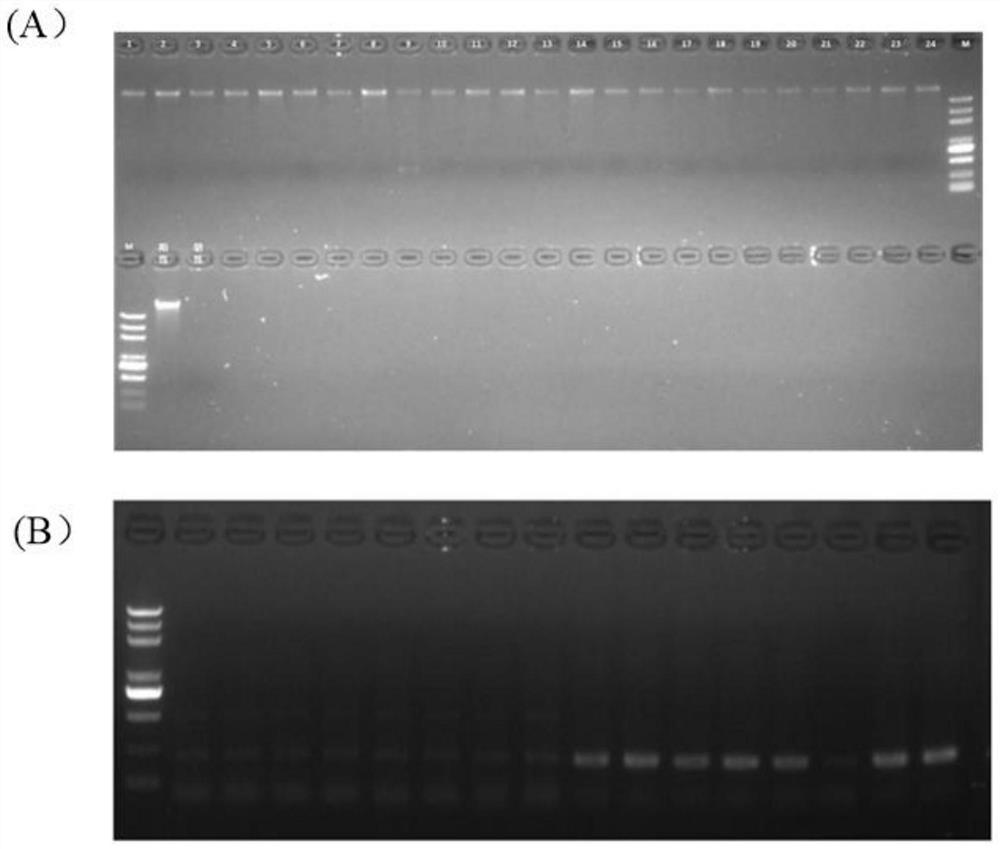
[0062] After stacking each material neatly, use scissors to cut an appropriate amount of leaves and place them in a 2mL centrifuge tube. Freeze the centrifuge tubes of the collected samples, use a tissue grinder to fully grind the leaves into powder, and use the Tiangen Plant Genomic DNA Extraction Kit to extract the genomic DNA of the sample to be tested. The concentration of DNA was detected with Nanodrop one, and the DNA was diluted to 10ng / μL with ultrapure water for later use. The sample extraction genomic DNA electrophore...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com