Polypeptiomics identification method based on Bayesian evaluation and sequence library searching and application thereof
An identification method and sequence technology, applied in the field of protein secondary mass spectrometry identification, can solve the problems of short peptide identification results, time-consuming, inability to identify short peptides, etc., and achieve the effect of reducing workload, improving efficiency, and improving identification efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
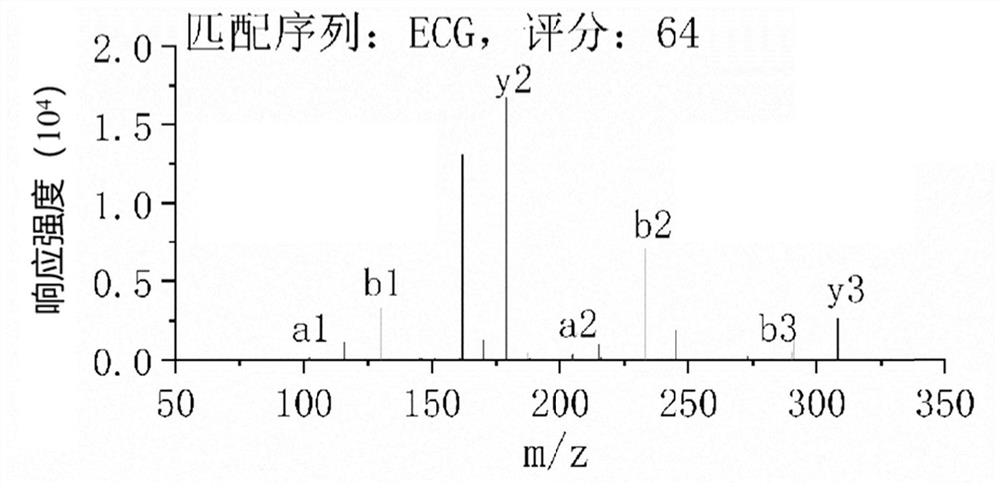
[0133] The analysis object is the mass spectrometry detection data collected by the high-resolution liquid-mass spectrometer to detect the glutathione standard (amino acid sequence: ECG). The analysis method is the same as the above-mentioned embodiment. The specific detection parameters are set as follows:
[0134] The protein sequence library is the soybean protein sequence library (downloaded from UniProt, searched for the keyword "soybean"). In step (2), the preset residue fragment length range is 1-3; in step (4), the threshold value of the response intensity of the primary precursor ion signal is 1000; in step (5), the threshold value of the coverage ratio of the secondary product ion is 1000. 70%; in step (6), the enzyme used in the simulated enzymatic hydrolysis is Alcalase broad-spectrum alkaline protease, and there is no restriction on the restriction site, and the preset length range of the polypeptide fragment to be retrieved is 2-10; step (8) Among them, the pri...
Embodiment 2
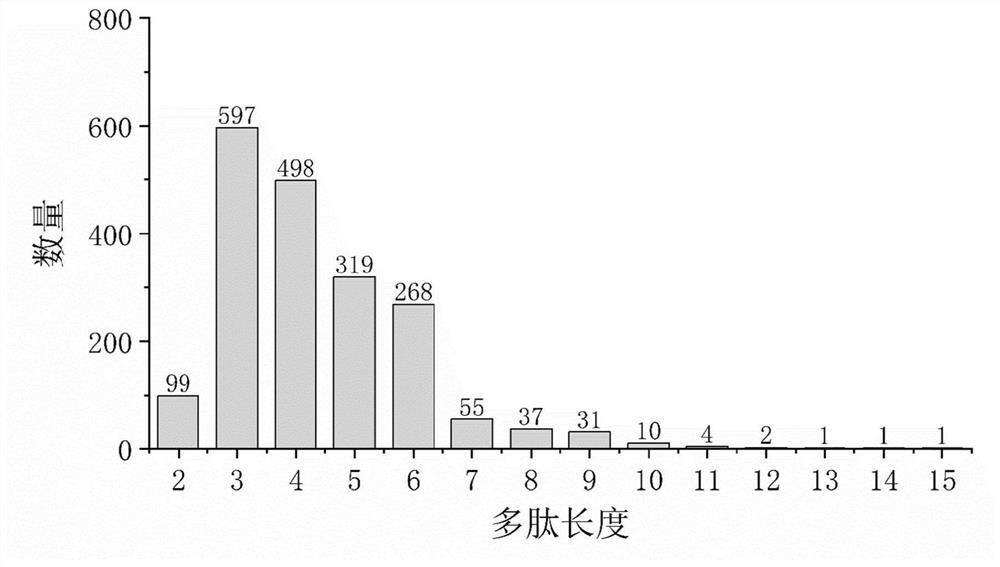
[0136] The object of analysis is the mass spectrometry detection data collected by high-resolution liquid-mass spectrometry to detect the enzymatic hydrolysis products of soybean protein (soybean protein is provided by Linyi Shansong Biological Products Co., Ltd., and the enzyme used is Alcalase broad-spectrum alkaline protease). . The analysis method is the same as the above-mentioned embodiment. The specific detection parameters are set as follows:
[0137] The protein sequence library is the soybean protein sequence library (downloaded from UniProt, searched for the keyword "soybean"). In step (2), the preset residue fragment length range is 1-3; in step (4), the threshold value of the response intensity of the primary precursor ion signal is 1000; in step (5), the threshold value of the coverage ratio of the secondary product ion is 1000. 30%; in step (6), the enzyme used in the simulated enzymatic hydrolysis is Alcalase broad-spectrum alkaline protease, and there is no ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com