Tumor specific promoter and application thereof
A promoter and promoter element technology, applied in the field of tumor-specific promoters, can solve problems such as difficult to obtain good results and limited activity, and achieve the effect of inhibiting tumor growth and improving the function of tumor-specific promoters
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0073] Example 1 Detection of mRNA expression of human telomerase reverse transcriptase in different human cells
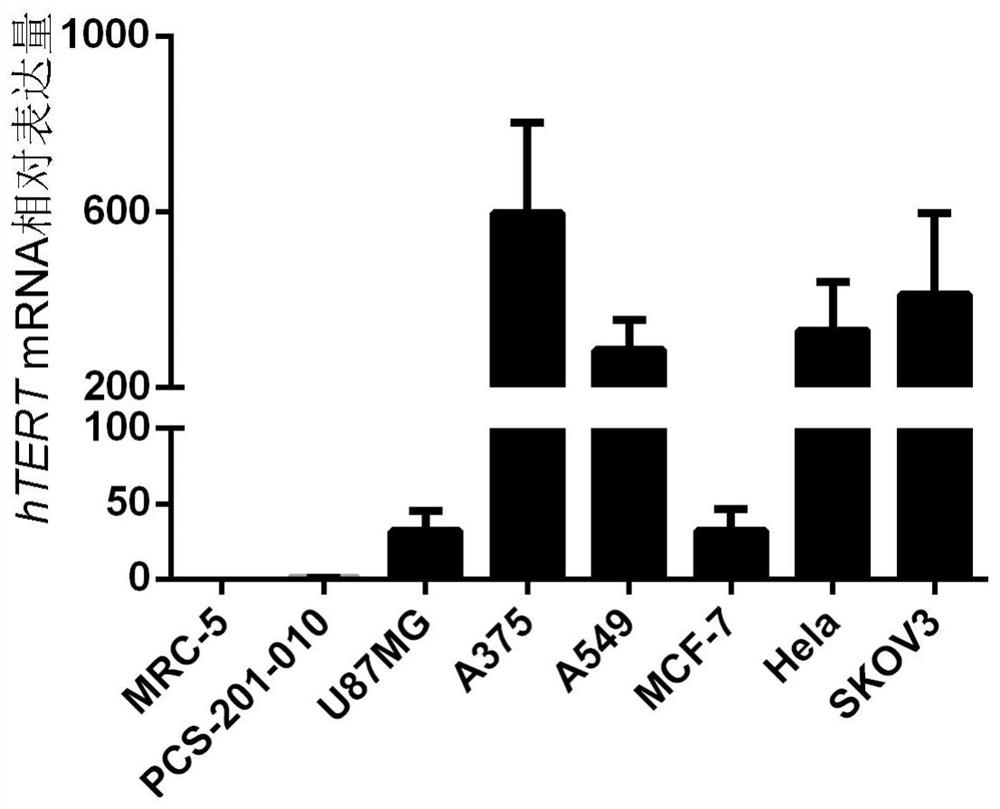
[0074] The cells used in this example are human-derived normal cells, human embryonic lung fibroblasts MRC-5 (ATCC: CCL-171) and human primary skin fibroblasts PCS-201-010 (ATCC: PCS-201-010 TM), human tumor cells: malignant glioblastoma cells U87MG (ATCC: HTB-14), malignant melanoma cells A375 (ATCC: CRL-1619), lung cancer cells A549 (ATCC: CRM-CCL-185), breast cancer cells Cells MCF-7 (ATCC: HTB-22), cervical cancer cells Hela (ATCC: CRM-CCL-2) and ovarian cancer cells SKOV3 (ATCC: HTB-77). Cells were harvested, and total cellular RNA was extracted and reverse transcribed into cDNA. The expression of human telomerase reverse transcriptase was detected by RT-PCR, and GAPDH was used as an internal reference.
[0075] The result is shown ( figure 1 ), in normal cells MRC-5 and PCS-201-010, human telomerase reverse transcriptase was almost not expressed, while in ...
Embodiment 2
[0076] Example 2 Detection of hTERT promoter mutation in different human tumor cells
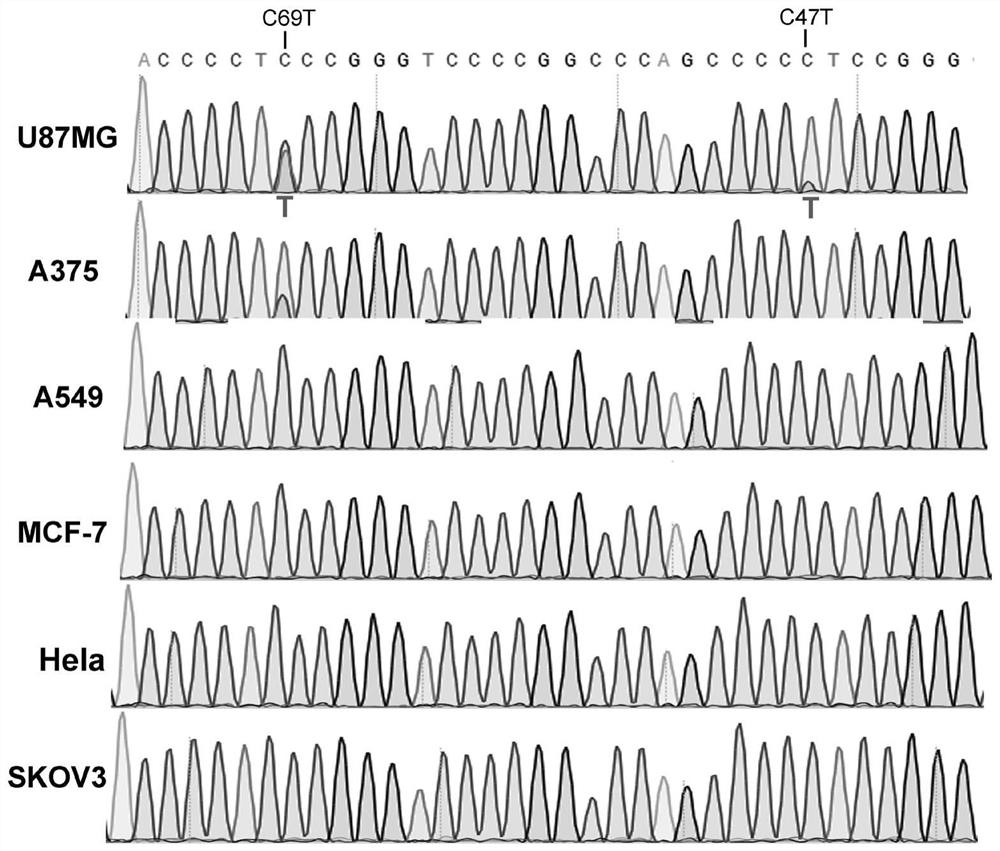
[0077] The tumor cells U87MG, A375, A549, MCF-7, Hela and SKOV3 were collected, the cell genome was extracted, and the hTERT promoter nucleic acid sequence was sequenced.
[0078] experiment result shows( figure 2 ): In U87MG cells, there are partial C69T and C47T double mutations in the hTERT promoter; in A375 cells, there is a C69T mutation; in other cells, there is no mutation in the hTERT promoter. Therefore, it was considered whether these mutations could be applied to the promoter optimization of oncolytic adenovirus and the improvement of oncolytic effect.
Embodiment 3
[0079] Example 3 Preliminary optimization of hTERT promoter
[0080] 1. Construction of hTERT promoter point mutation
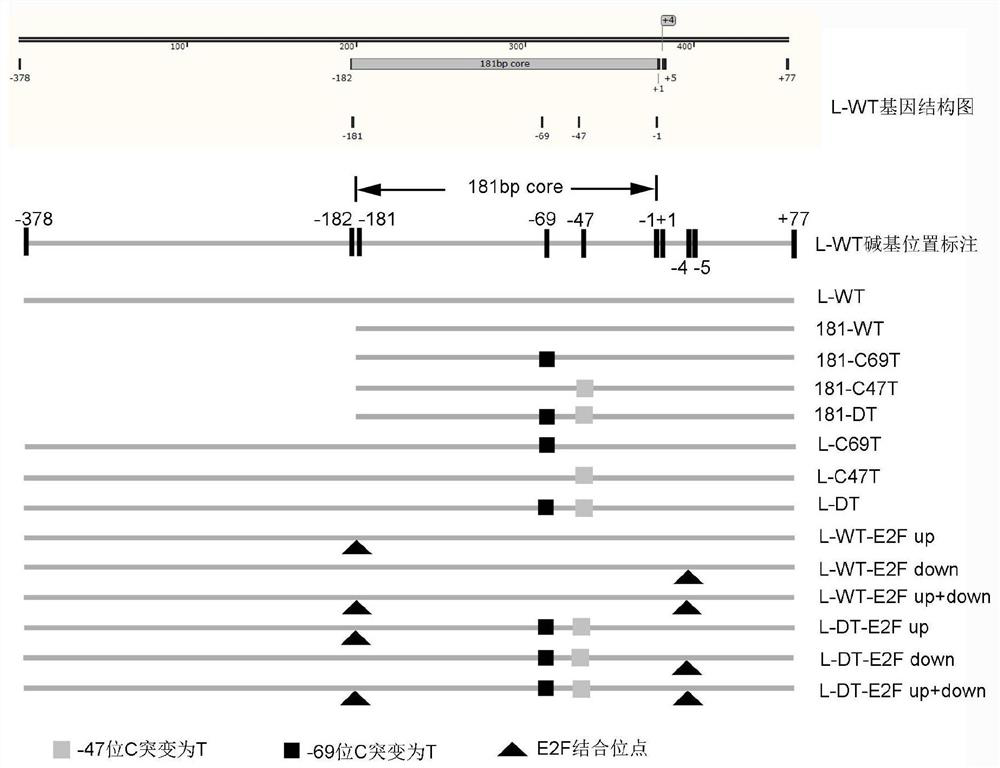
[0081] This example uses the -378-+77 region sequence of the hTERT promoter (from the TERT 5' regulatory region of human chromosome 5, sequence ID: NG_055467.1), a total of 455 bp, marked as hTERT WT ( image 3 The winning bid is L-WT) sequence (SEQ ID No. 2) (see image 3 ). By studying its sequence, the segment containing 181 bp from -181 to +77 was selected as the core region with strong promoter activity, which was marked as 181-WT sequence (SEQ ID No. 1) (see image 3 ). image 3 Where +1 represents the first nucleotide in the mRNA sequence, and -1 represents the first 5' nucleotide to the transcription start site. Both C69T (nucleotide C at -69) and C47T (nucleotide C at -47) indicate that the hTERT promoter sequence has a single cytosine C to thymine T mutation. DT indicated that the sequence had the above-mentioned double mutation of C→T at posit...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com