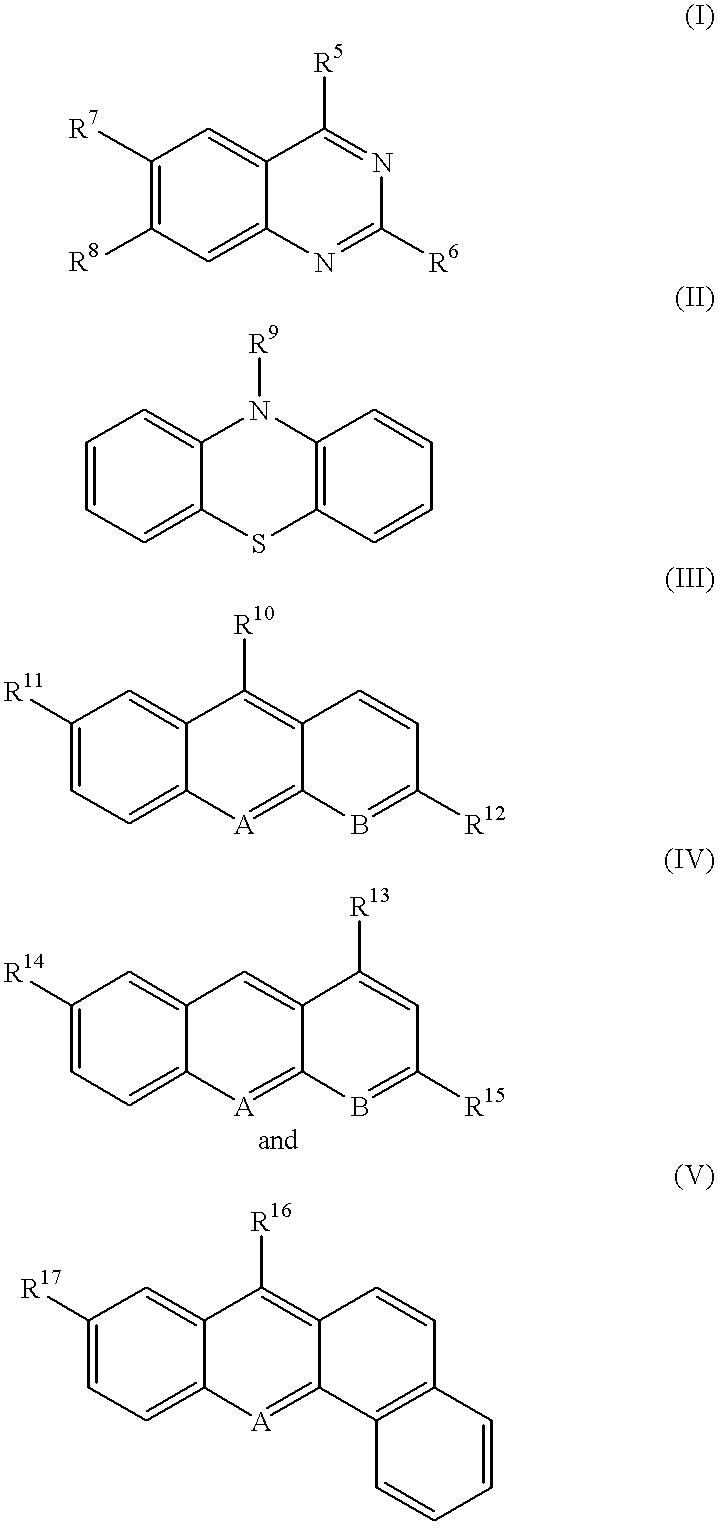
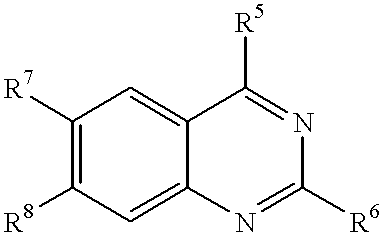
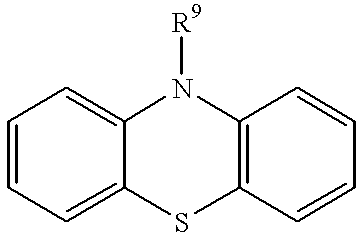
Methods and composition for restoring conformational stability of a protein of the p53 family
a technology of conformational stability and p53 family, which is applied in the field of cancer treatment, can solve the problems of less capable of retaining native conformation, uncontrolled proliferation of affected cells and tumor growth, and impaired cell function, and achieve negative dna binding effect, enhanced conformational stability, and wild-type activity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
VI. EXAMPLE 1
p53 DBD Thermostabilization Assay
[0186] A high through-put assay using wild-type p53 DBD was developed. Pharmacological compounds were screened using the assay, and those compounds that stabilized the active conformation of the DBD were scored as hits.
[0187] A. Materials and Methods
[0188] Thermostabilization Assay. Recombinant DBD (residues 94-312) from wild-tvpe and mutant p53 proteins and FLAG-tagged p53 DBD were prepared as described (Pavletich et al., 1993, Genes and Dev. 7, 2556-2564; Bullock et al., 1997, supra.). Mutant proteins used were 143A, 173A, 175S. 249S, and 273H. A number of small molecule organic compounds were tested. Compound stocks were dissolved in DMSO at 10 mg / ml and diluted prior to use. The proteins (0.25-1.0 ng / well) were diluted in a buffer containing 25 mM HEPES, pH 6.8, 150 mM KCl, 10 mM dithiothreitol and attached in 50 ul to Reacti-Bind microtiter plates (Pierce) for 35 minutes on ice. The wells were rinsed with 25 mM HEPES, pH 6.8, 150 mM...
example 2
VII. EXAMPLE 2
Determination of p53 Conformation in Cells and Tumors
[0198] In this example and the examples that follow, prototype compounds are shown to function at low micromolar concentrations to modulate mutant p53 in living cells and in tumors and to suppress the growth of tumors with naturally mutated p53.
[0199] A. Materials and Methods
[0200] Cell Culture. All cell lines were obtained from the ATCC and grown in the recommended media with 10 percent fetal calf serum (Gibco BRL).
[0201] Determination of p53 Conforniation Approximately 1.times.10.sup.7H1299 / Reporter+Mutant p53 cells were treated overnight, rinsed three times with cold Tris buffered saline, and lysed in 1.5 ml of hypotonic lysis buffer (20 mM HEPES, pH 7.4, 10 mM NaCl, 20 percent glycerol, 0.2 mM EDTA, 0.1 percent Triton-X 100, 10 mM dithiothreitol with protease inhibitors). Cells were pelleted in microfuge tubes at 2000 rpm for 5 minutes at 4.degree. C. and nuclear extracts were prepared by resuspending the pellet...
example 3
VIII. EXAMPLE 3
Restoration of p53 Function
[0206] A. Materials and Methods
[0207] Transactivation assays. Cells were transfected with expression plasmids encoding mutant p53 proteins (173A, 249S) and a neomycin selectable marker using DOTAP cationic lipid transfection-reagent (Boehringer Mannheim) or calcium phosphate. Cells were also transfected with a plasmid encoding the hygromycin resistance marker and a p53 reporter gene comprised of four copies of a p53 binding sequence corresponding to a p53 binding sequence in the promoter region of the Herpes Simplex virus thymidine kinase gene (base numbers 26 to 58 of GenBank accession no. S57428 thymidine kinase, which begins with he sequence GCCTTGCCT and ends with the sequence TGCCTTTTC) placed upstream of he SV40 basal promoter driving the luciferase gene. A matched cell pair was prepared by transfecting a clone of cells with the reporter construct with an additional construct for mutant p53 expression. Transfected clones were selected ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| length | aaaaa | aaaaa |
| length | aaaaa | aaaaa |
| length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com