Deep learning-based transcription factor binding site positioning method
A transcription factor and binding site technology, applied in the field of deep learning, can solve the problems of long response time, poor performance, and low efficiency, and achieve fast response time, good performance, and accurate prediction and positioning
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
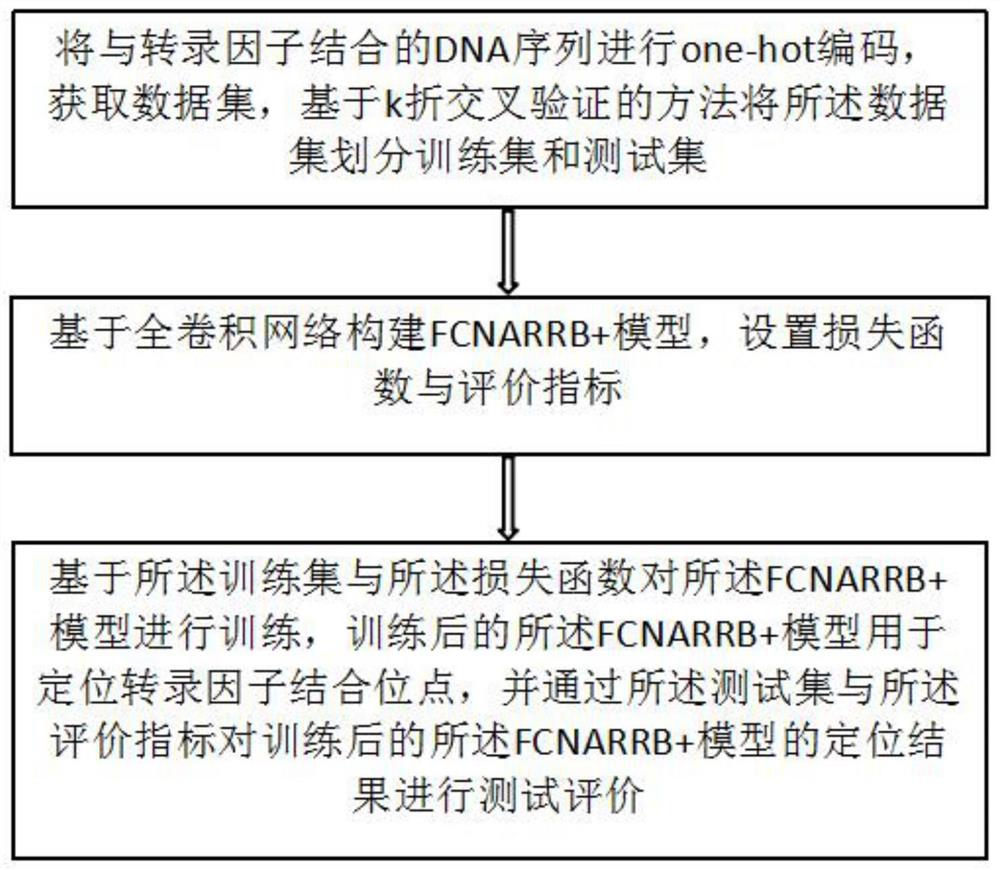
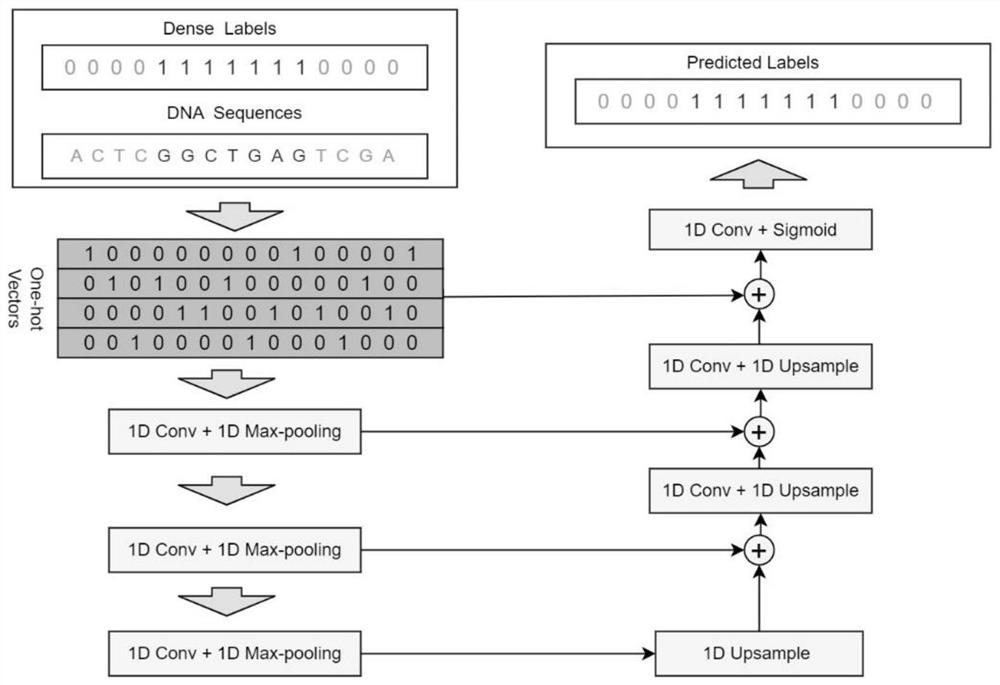
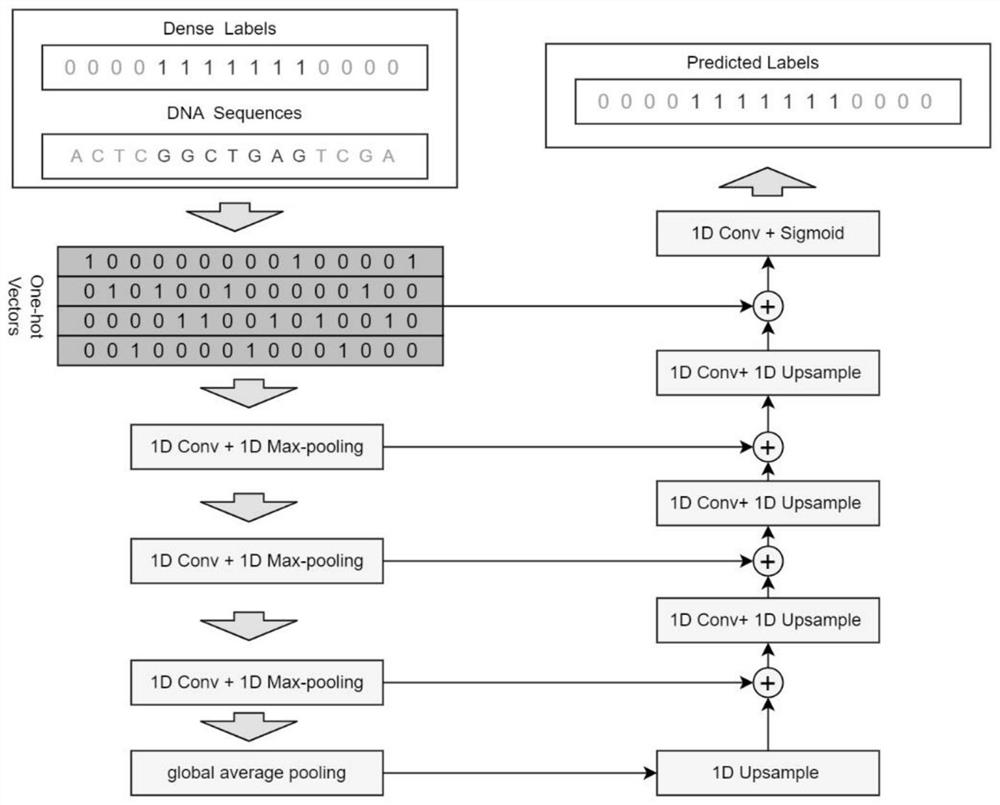
[0050] like figure 1 As shown, this embodiment provides a deep learning-based method for locating transcription factor binding sites, including:
[0051] One-hot encoding is performed on the DNA sequence bound to the transcription factor to obtain a data set, and the data set is divided into a training set and a test set based on the k-fold cross-validation method;
[0052] Further, one-hot encoding is performed on the DNA sequence bound to the transcription factor: data encoding is performed on the bases {A, C, G, T} in the DNA sequence according to one-hot encoding, and the DNA sequence is encoded by one-hot encoding. The conserved information data in the sequence is selected from the data at the corresponding position, and the two constitute the coding information of the DNA sequence.
[0053] In this embodiment, a given DNA sequence (assuming a length of L) is one-hot encoded as input data, and at the same time, a Dense Label label (length is L, and the element value of t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com