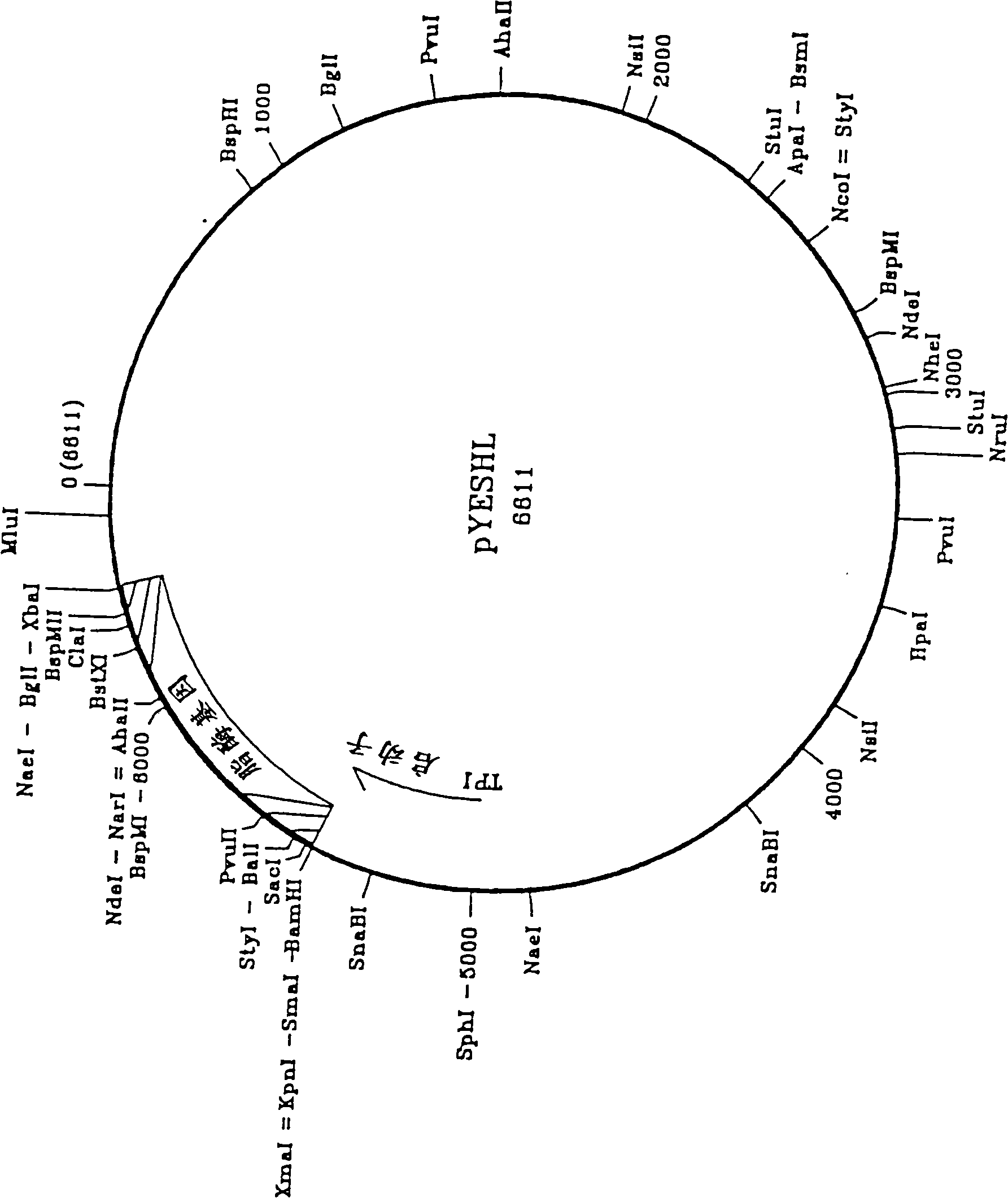
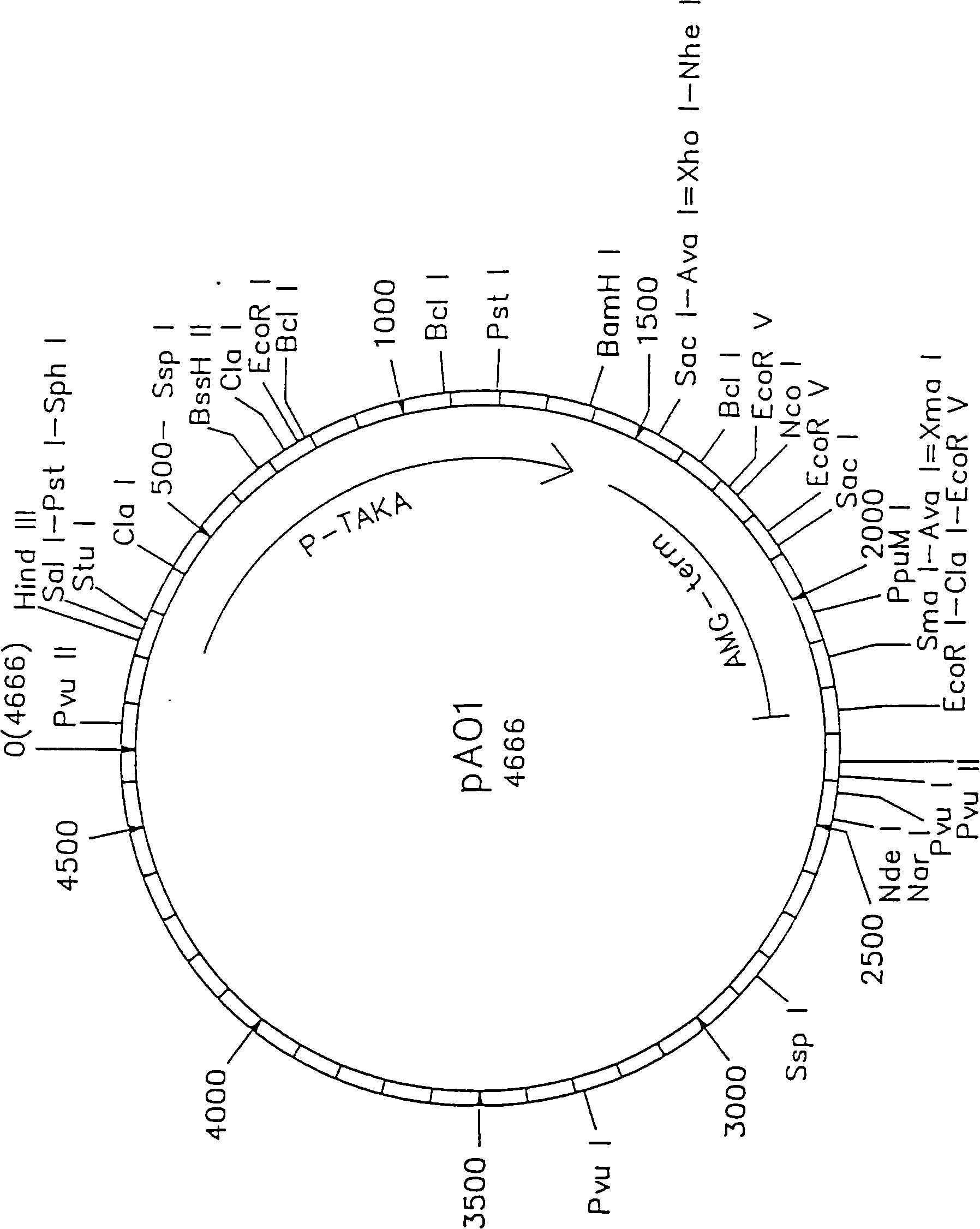
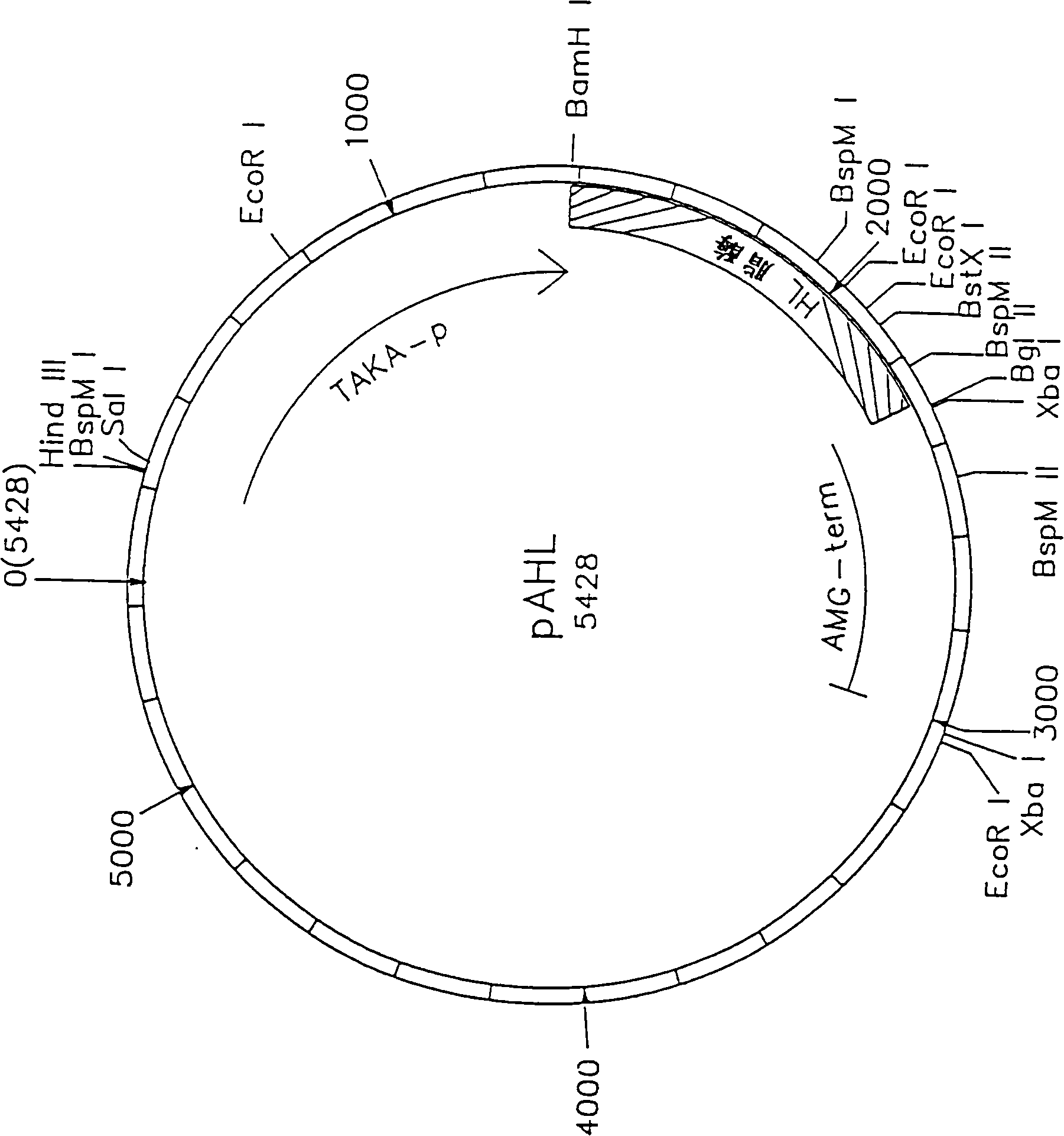
Novel lipolytic enzymes
A lipolytic enzyme, lard technology, applied in the field of lipolytic enzymes, can solve the problem of no fat dirt and high activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0516] The preparation of homologous DNA fragments in step c) can be carried out by amplifying homologous DNA sequences (e.g. comprising one or more mutations in the lipolytic gene and contained in a plasmid or vector) using any suitable method, so A suitable method is such as the standard PCR amplification method described in US Patent 4,683,202 or Saiki et al., (1988), Science, 239, 487-491.
[0517] The vector can be introduced into the recombinant host cell by transformation (step d). Where the recombinant host cell is a S. cerevisiae strain such as S. cerevisiae YNG318 (described below), transformation can be performed as described in Sambrooks et al. ((1989), Molecular Cloning: A Laboratory Manual, Second Edition, Cold Spring Harbor, NY, USA ) proceed as described.
[0518] Screening for positive lipolytic enzyme variants can be performed, for example, by the screening methods described above for random mutagenesis.
[0519] One of the cycles of steps a) to f) may be p...
Embodiment 1
[1215] Construction of random lipolytic enzyme variants
[1216] Random mutagenesis libraries of the complete H. lanuginosa lipolytic enzyme gene and its amino acids (aa) 91-97 and 206-211 were prepared as described above in Materials and Methods.
[1217] Amino acid regions 91-97 and 206-211 were selected for the first round of domain-specific mutagenesis because these regions have been found to be important for wash performance. Regions 91-97 are part of the lid region of the enzyme and regions 206-211 form part of the hydrophobic cleft of the enzyme.
[1218] An oligonucleotide containing 93% of the wild-type nucleotide and 2.33% each of the other three nucleotides at the amino acid codon to be mutagenized was synthesized for each region. Where no amino acid was likely to be changed, the third nucleotide in the codon (the wobble base) was synthesized with 50% G / 50% C to give a greater probability of changing to an amino acid containing one or two codons. The composition o...
Embodiment 2
[1456] Construction of a first wash variant of H. lanuginosa lipolytic enzyme
[1457] 1. Domain Shuffling by Recombination and Screening
[1458] Twenty H. lanuginosa lipolytic enzyme variants, some of which were constructed according to Example 1, with good wash performance (as assessed in various wash-related assays) were available as described herein in the Materials and Methods section. The method of in vivo recombination in Saccharomyces cerevisiae YNG318 is used for recombination. The lipolytic enzyme variants utilized are evident from Table 1. Most of these variants were selected for reduced calcium dependence and increased tolerance to the detergent component Dobanol 25-7 (see Materials and Methods section above) by random or localized random mutagenesis as described in the Materials and Methods section above. And build. Some variants are the result of two or more consecutive cycles of mutagenesis and selection.
[1459] The restriction enzyme-opened vector and th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com